Quick Links
Labs
Selected Publications
Spatial transcriptomics reveals altered communities and drivers of aberrant epithelia and pro-fibrotic fibroblasts in interstitial lung diseases.
Cell Genom. 2026;:101066
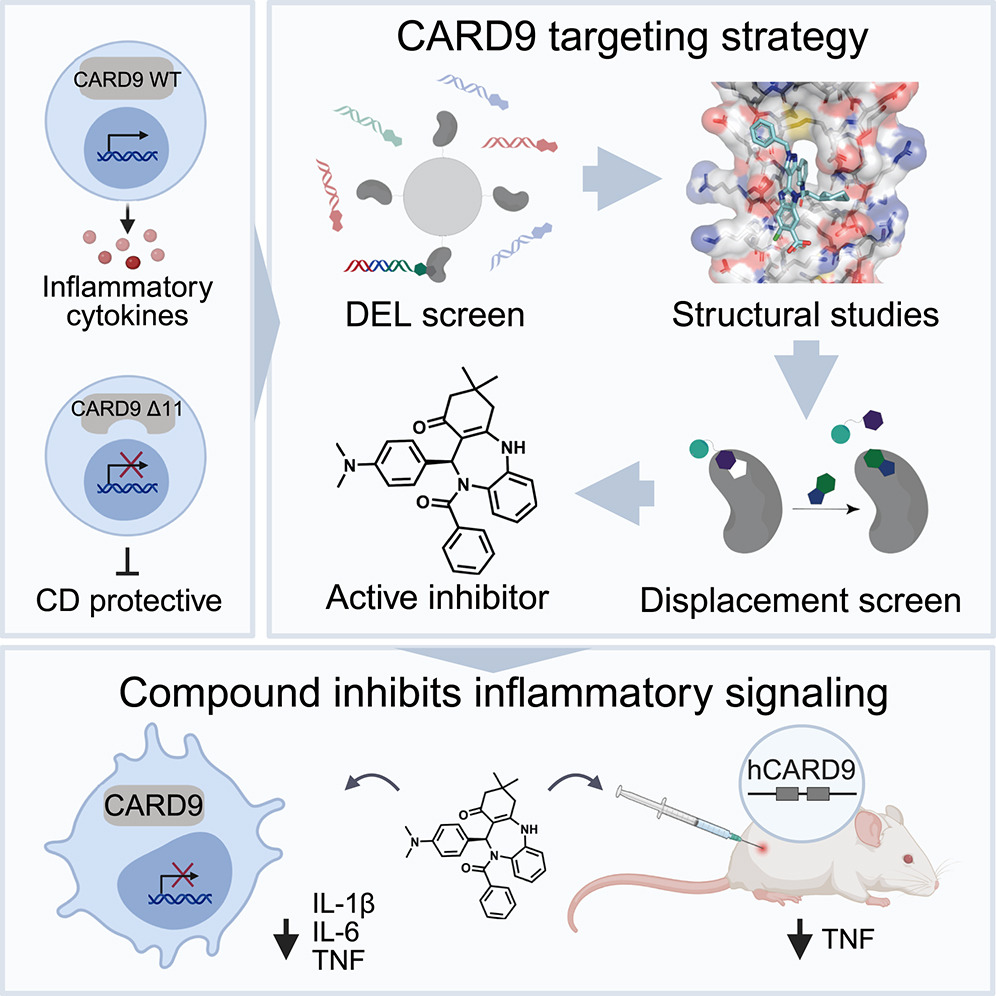
Human genetics guides the discovery of CARD9 inhibitors with anti-inflammatory activity.
Cell. 2026;:ePub
Bridging AI and biology: Foundation models meet human physiology and disease.
Med. 2026;7(1):100971
Bidirectional CRISPR screens decode a GLIS3-dependent fibrotic cell circuit.
Nature. 2026;650(8103):997-1006
Protein language models uncover carbohydrate-active enzyme function in metagenomics.
BMC Bioinformatics. 2025;26(1):285
Regional encoding of enteric nervous system responses to microbiota and type 2 inflammation.
Science. 2025;390(6772):eadr3545
Human immunodeficiency virus and antiretroviral therapies exert distinct influences across diverse gut microbiomes.
Nat Microbiol. 2025;10(11):2720-2735
Nuclear receptor coregulator NRIP1 R448G modulates T cell gut homing to control intestinal inflammation.
Proc Natl Acad Sci U S A. 2025;122(38):e2508269122
Autoimmune disease risk gene ANKRD55 promotes TH17 effector function through metabolic modulation.
J Exp Med. 2025;222(11):ePub
Single-cell and spatial transcriptomics of stricturing Crohn's disease highlights a fibrosis-associated network.
Nat Genet. 2025;57(7):1742-1753
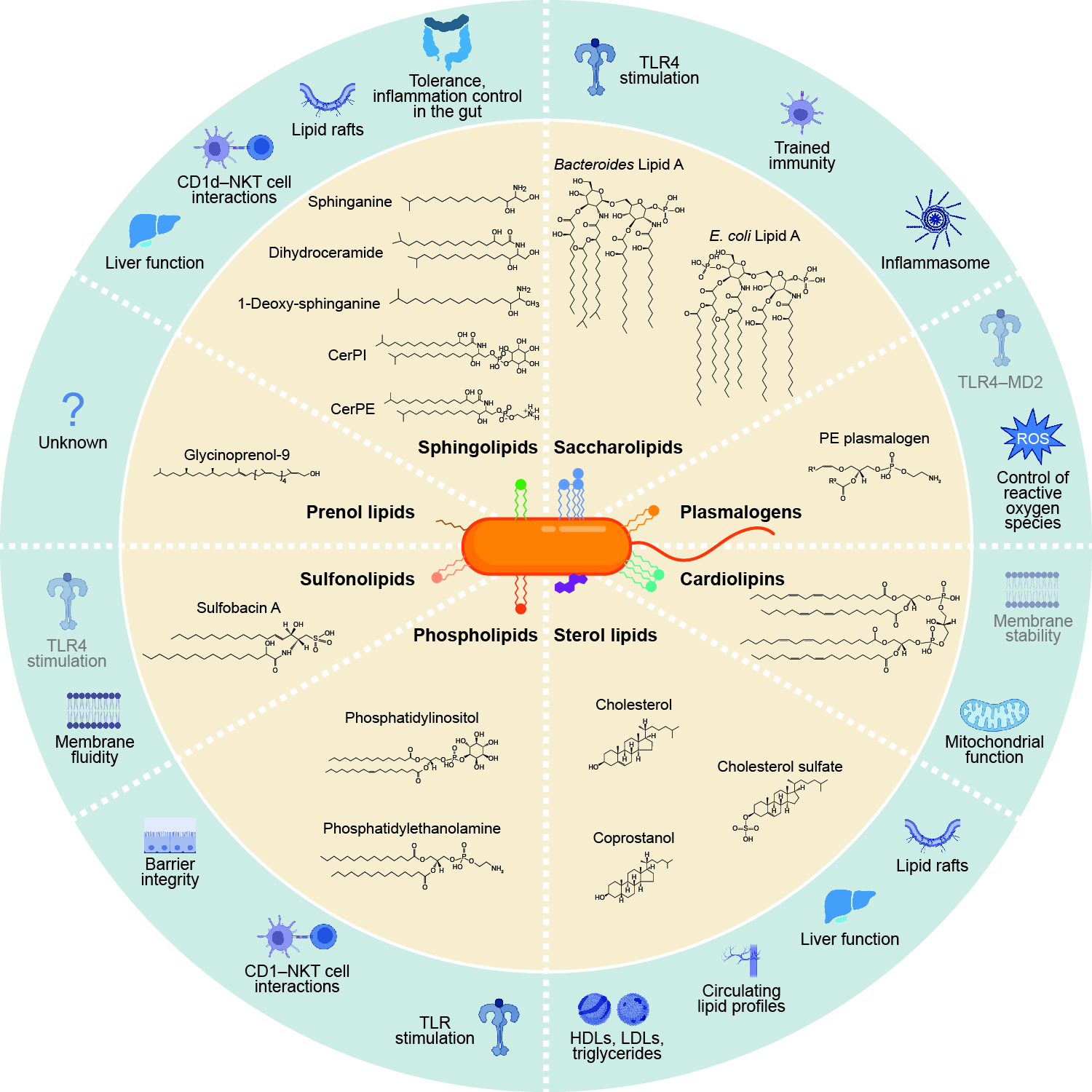
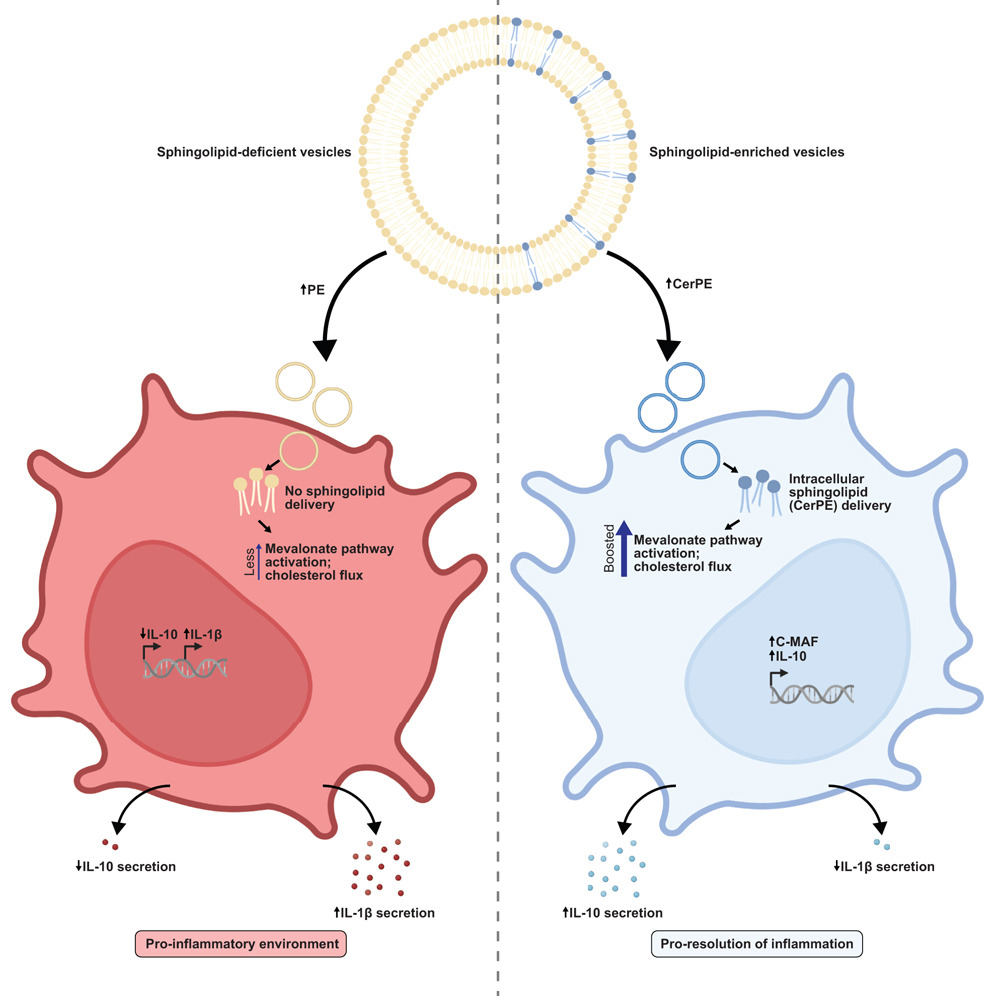
Bacteroides sphingolipids promote anti-inflammatory responses through the mevalonate pathway.
Cell Host Microbe. 2025;33(6):901-914.e6
Type 2 cytokines act on enteric sensory neurons to regulate neuropeptide-driven host defense.
Science. 2025;389(6757):260-267
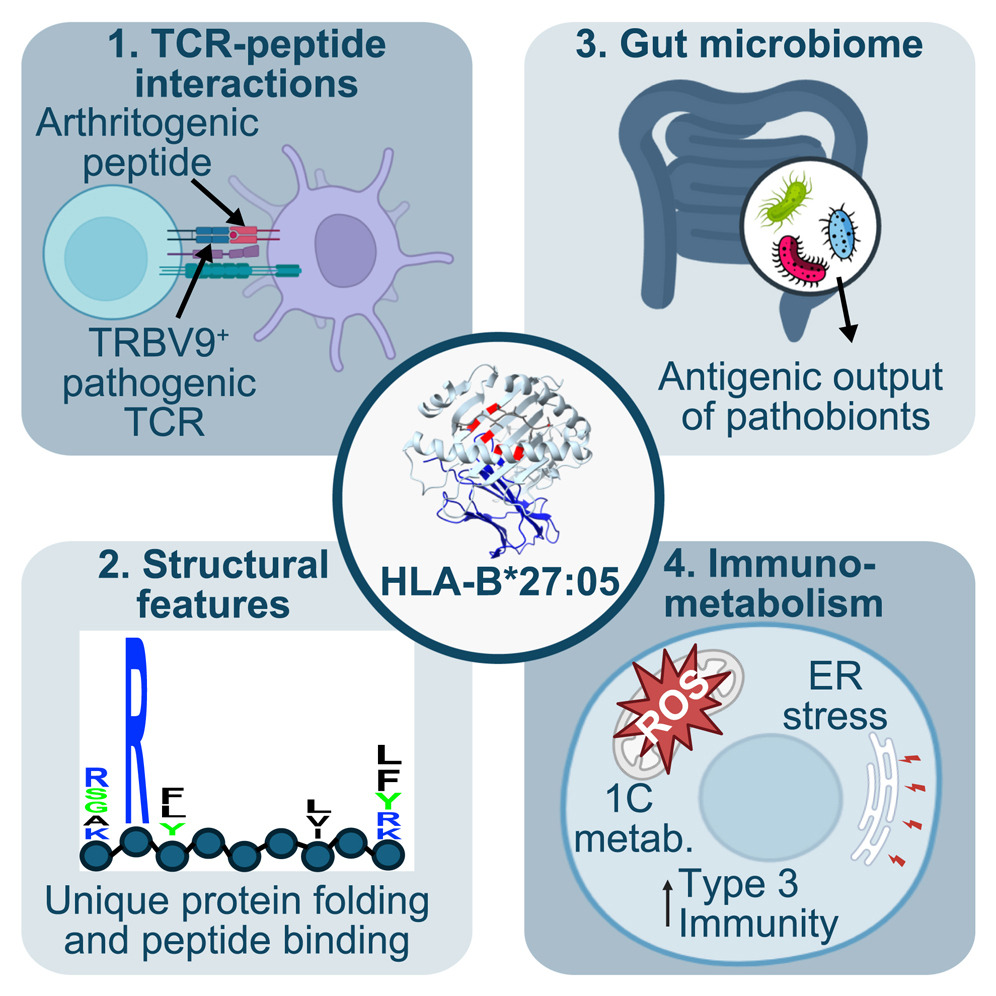
Emerging biochemical, microbial and immunological evidence in the search for why HLA-B∗27 confers risk for spondyloarthritis.
Cell Chem Biol. 2025;32(1):12-24
Unusual Phospholipids from Morganella morganii Linked to Depression.
J Am Chem Soc. 2025;147(4):2998-3002
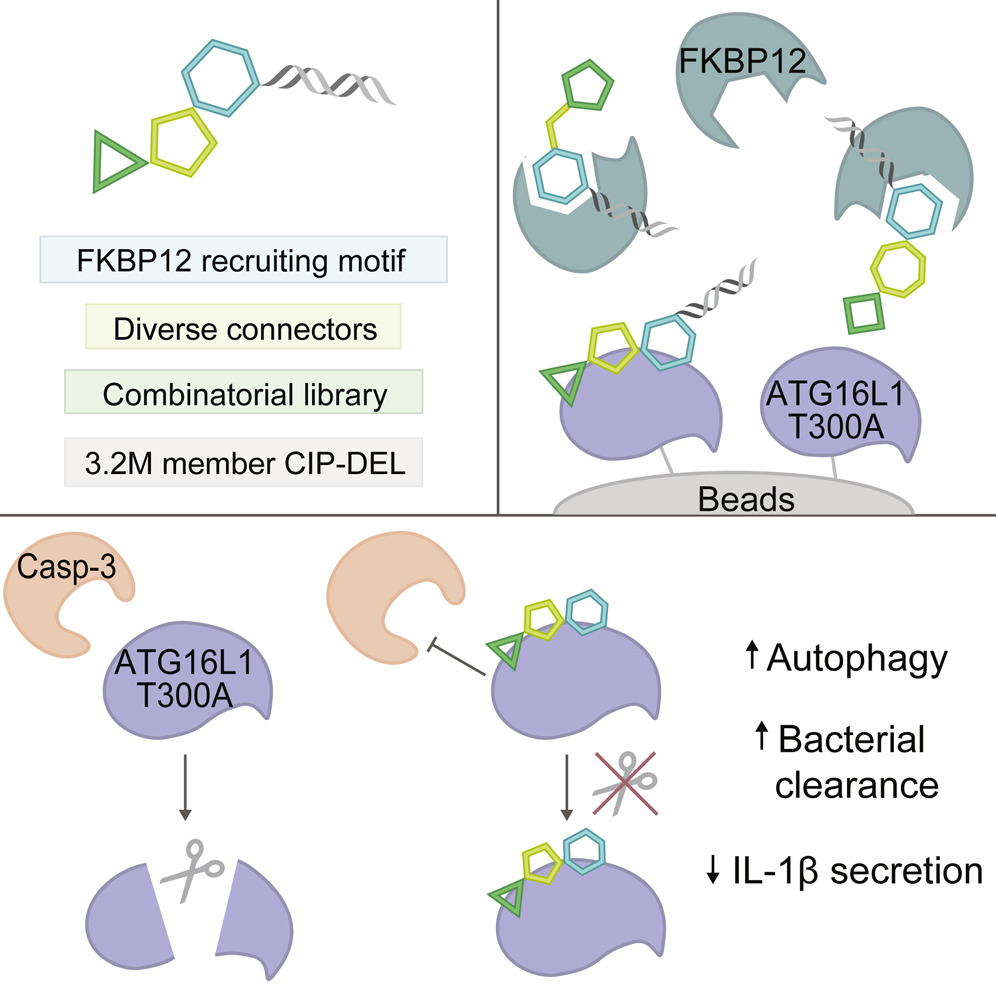
Development of an FKBP12-recruiting chemical-induced proximity DNA-encoded library and its application to discover an autophagy potentiator.
Cell Chem Biol. 2025;32(3):498-510.e35
Spatially restricted immune and microbiota-driven adaptation of the gut.
Nature. 2024;636(8042):447-456
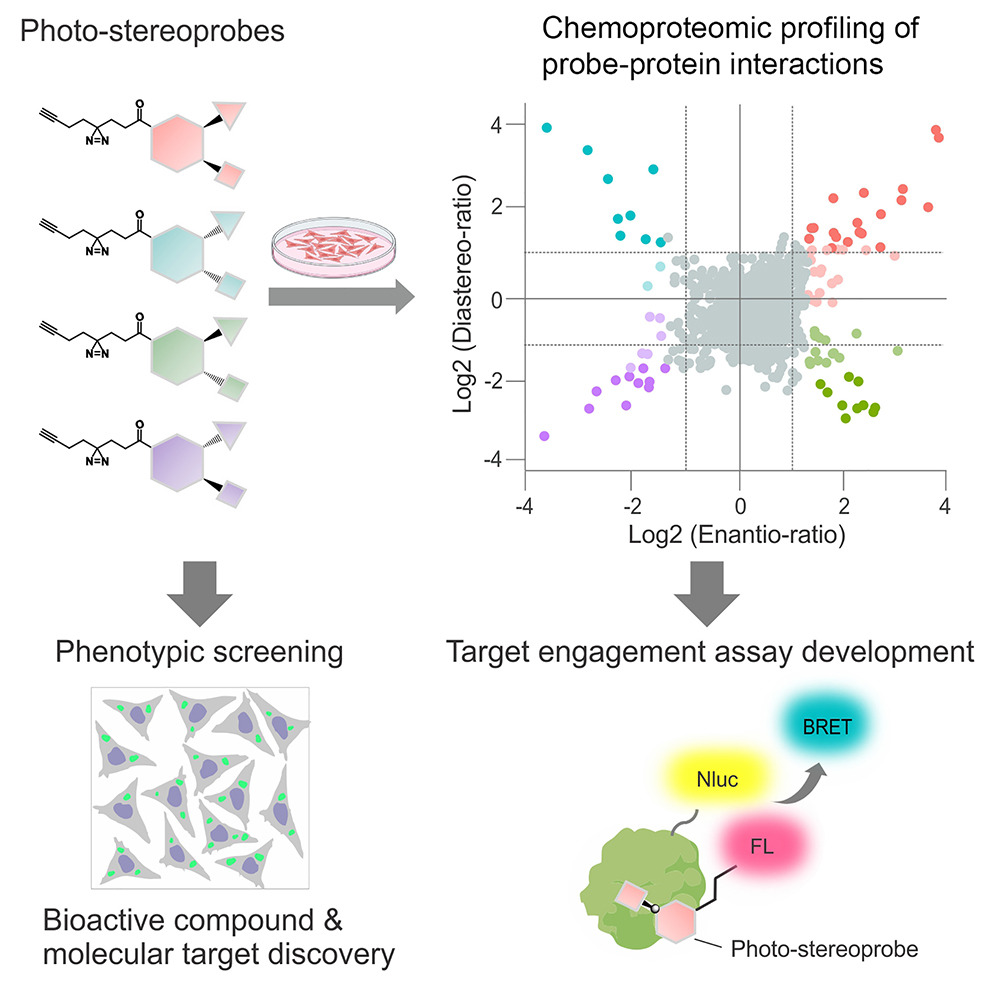
Chemical tools to expand the ligandable proteome: Diversity-oriented synthesis-based photoreactive stereoprobes.
Cell Chem Biol. 2024;31(12):2138-2155.e32
Commensal consortia decolonize Enterobacteriaceae via ecological control.
Nature. 2024;633(8031):878-886
B cell tolerance and autoimmunity: Lessons from repertoires.
J Exp Med. 2024;221(9):ePub
Small-molecule probe for IBD risk variant GPR65 I231L alters cytokine signaling networks through positive allosteric modulation.
Sci Adv. 2024;10(29):eadn2339
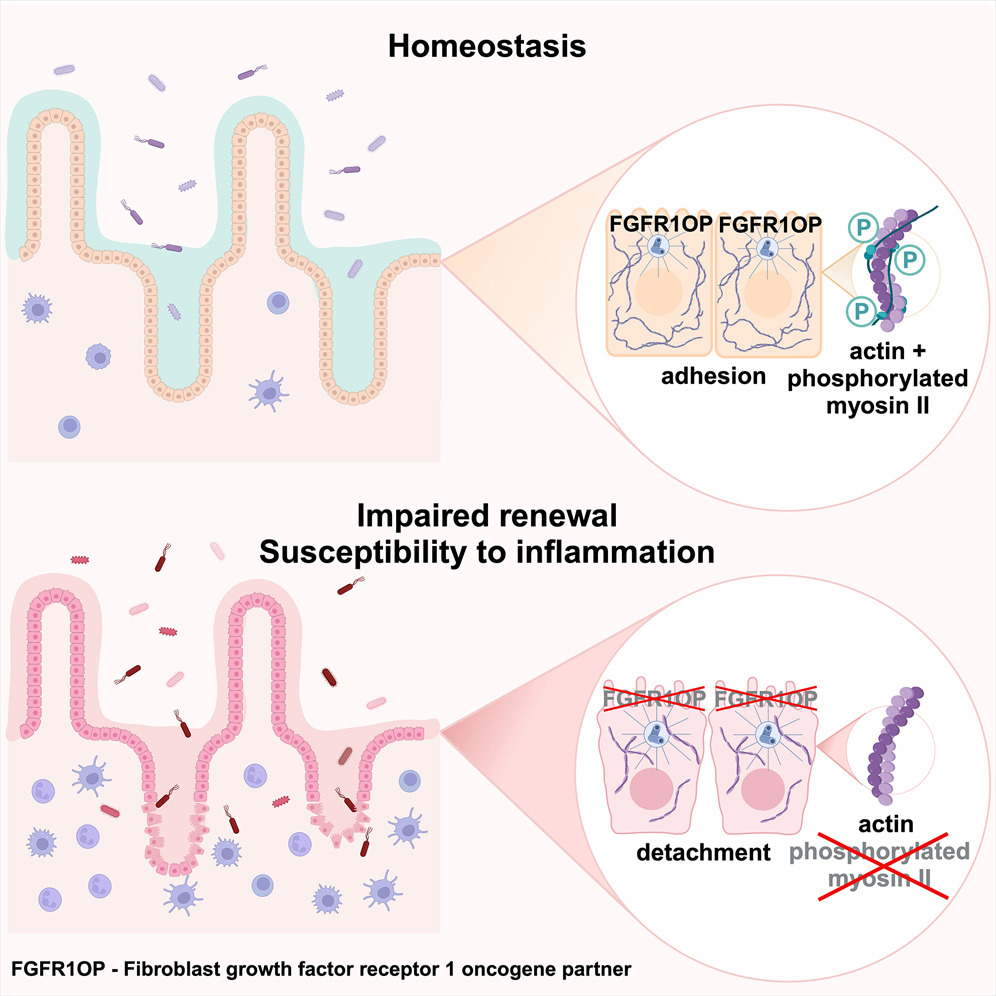
The centrosomal protein FGFR1OP controls myosin function in murine intestinal epithelial cells.
Dev Cell. 2024;59(18):2460-2476.e10
An esophagus cell atlas reveals dynamic rewiring during active eosinophilic esophagitis and remission.
Nat Commun. 2024;15(1):3344
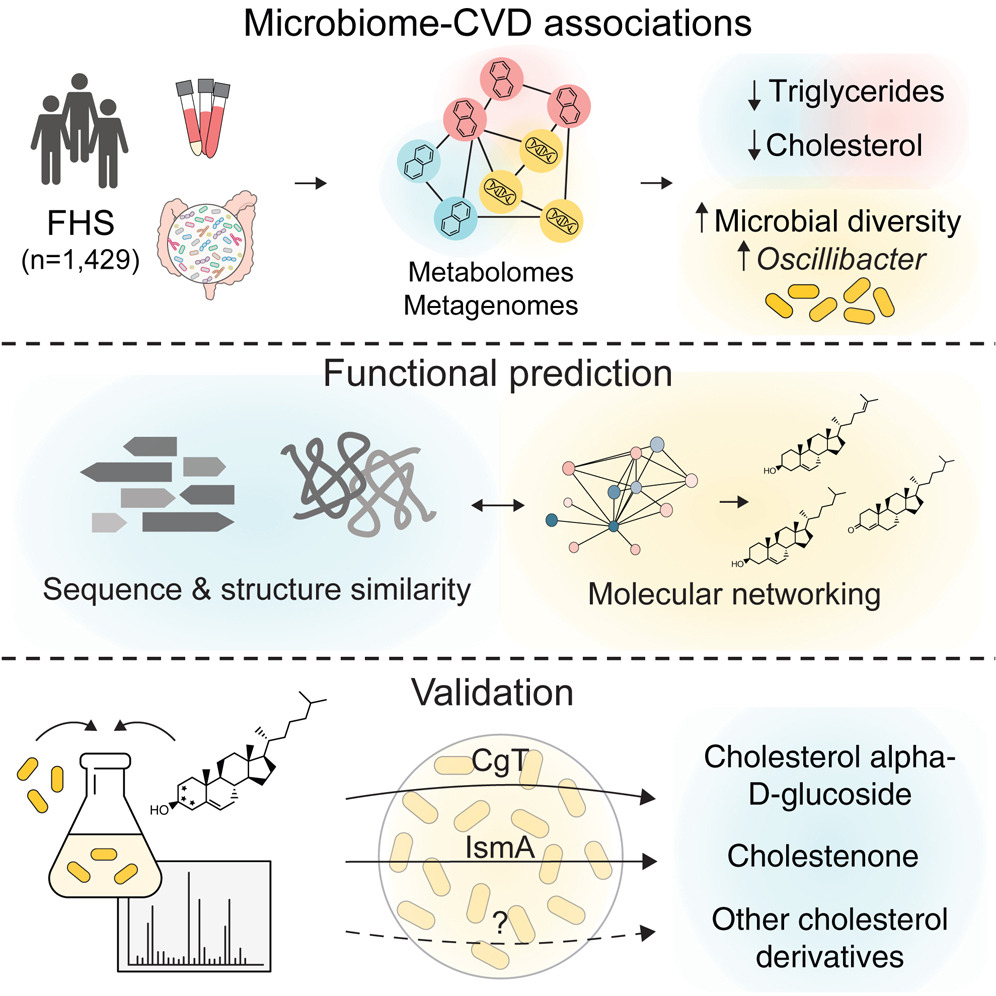
Gut microbiome and metabolome profiling in Framingham heart study reveals cholesterol-metabolizing bacteria.
Cell. 2024;187(8):1834-1852.e19
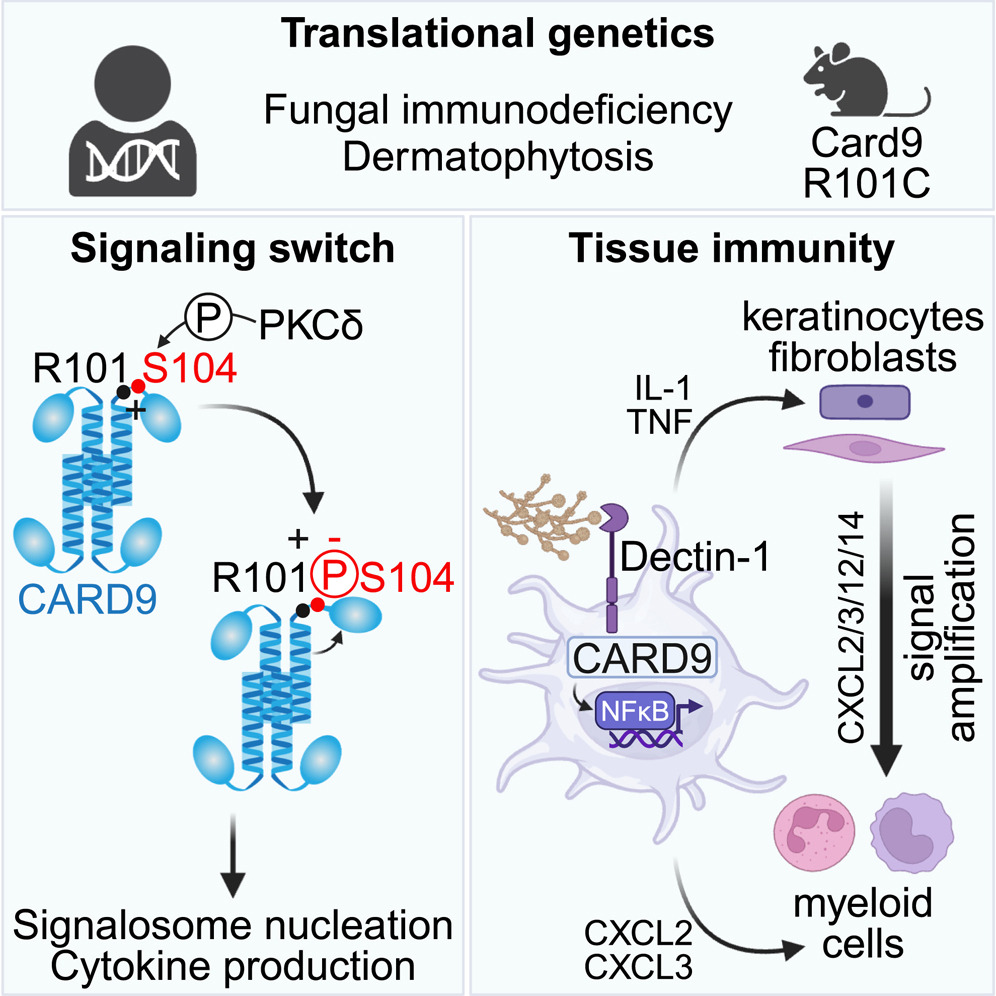
Translational genetics identifies a phosphorylation switch in CARD9 required for innate inflammatory responses.
Cell Rep. 2024;43(3):113944
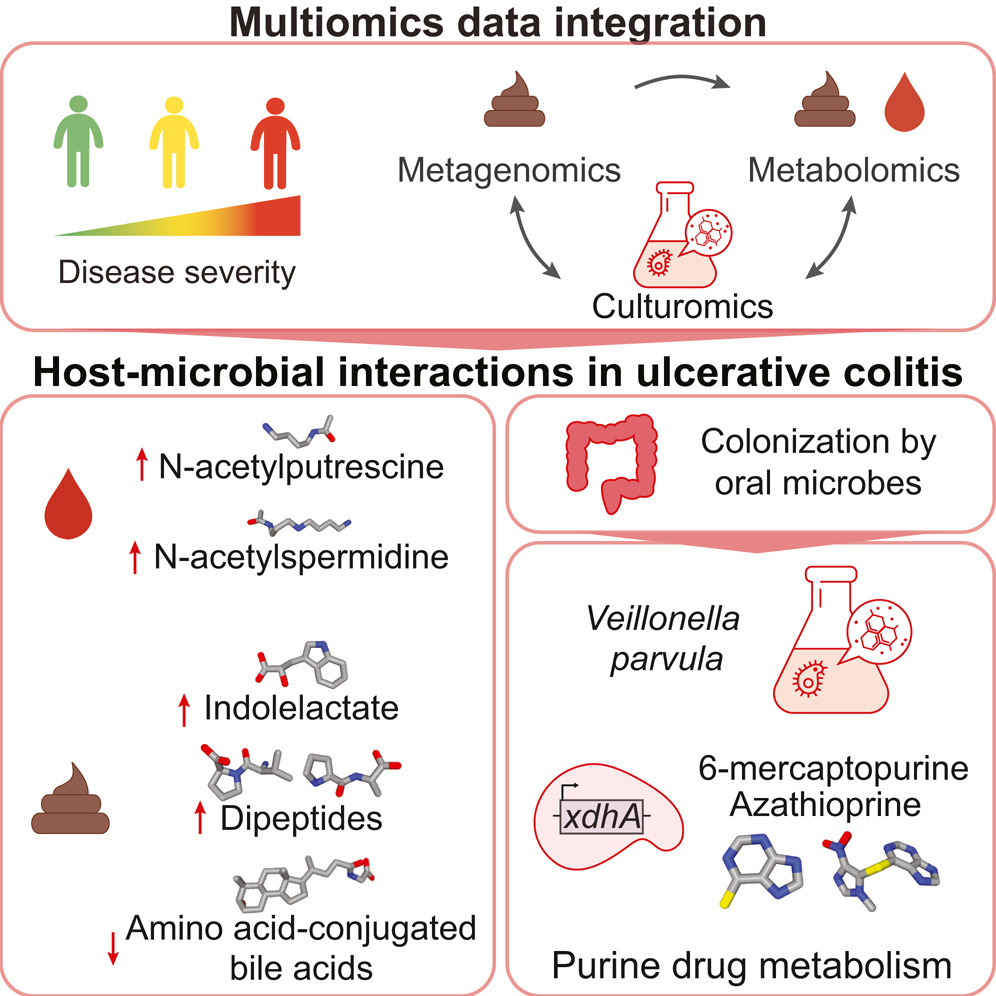
Linking microbial genes to plasma and stool metabolites uncovers host-microbial interactions underlying ulcerative colitis disease course.
Cell Host Microbe. 2024;32(2):209-226.e7
Host genetic regulation of human gut microbial structural variation.
Nature. 2024;625(7996):813-821
Identification of highly selective SIK1/2 inhibitors that modulate innate immune activation and suppress intestinal inflammation.
Proc Natl Acad Sci U S A. 2023;121(1):e2307086120
Reverse metabolomics for the discovery of chemical structures from humans.
Nature. 2023;626(7998):419-426
Multi-modal skin atlas identifies a multicellular immune-stromal community associated with altered cornification and specific T cell expansion in atopic dermatitis.
bioRxiv. 2023;:ePub
Genetic vulnerability to Crohn's disease reveals a spatially resolved epithelial restitution program.
Sci Transl Med. 2023;15(719):eadg5252
Protein Language Models Uncover Carbohydrate-Active Enzyme Function in Metagenomics.
bioRxiv. 2023;:ePub
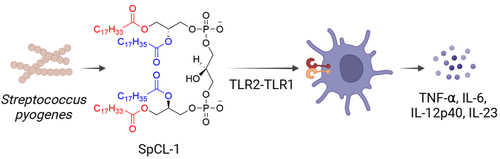
Revisiting Coley's Toxins: Immunogenic Cardiolipins from Streptococcus pyogenes.
J Am Chem Soc. 2023;145(39):21183-21188
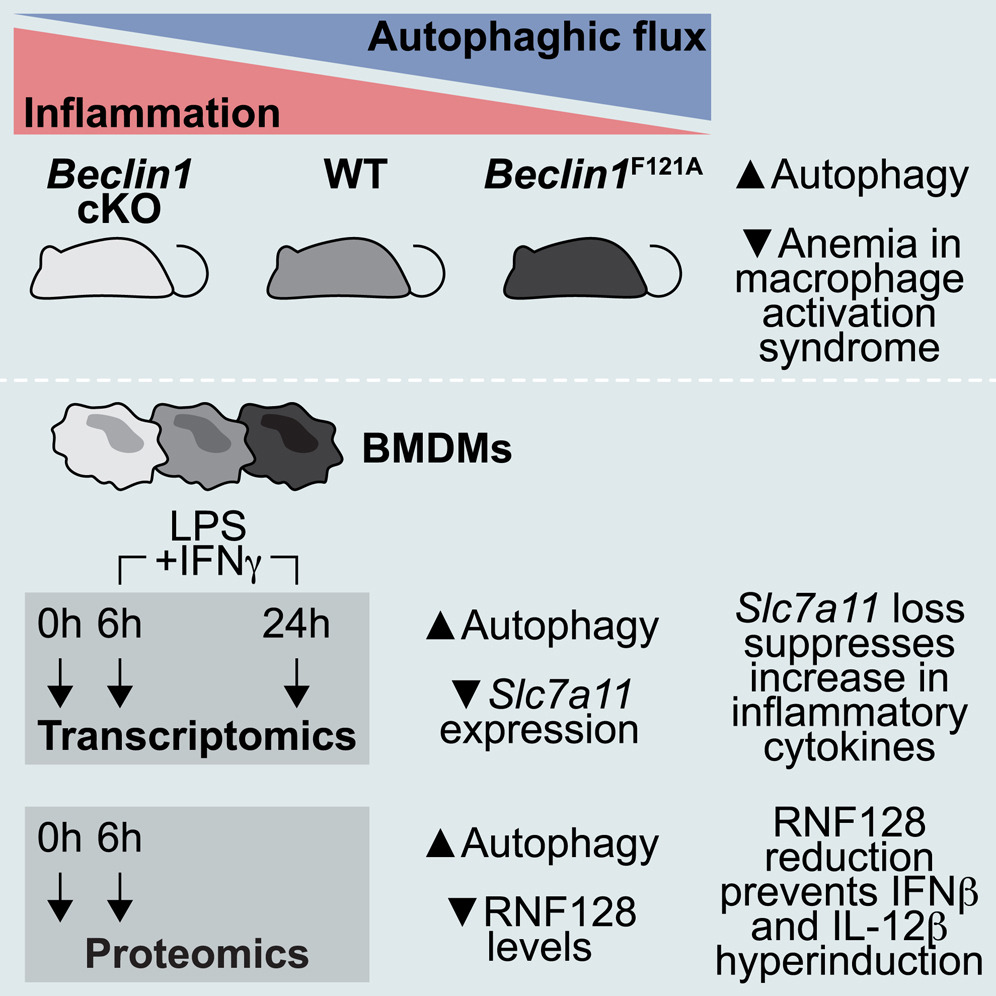
Constitutively active autophagy in macrophages dampens inflammation through metabolic and post-transcriptional regulation of cytokine production.
Cell Rep. 2023;42(7):112708
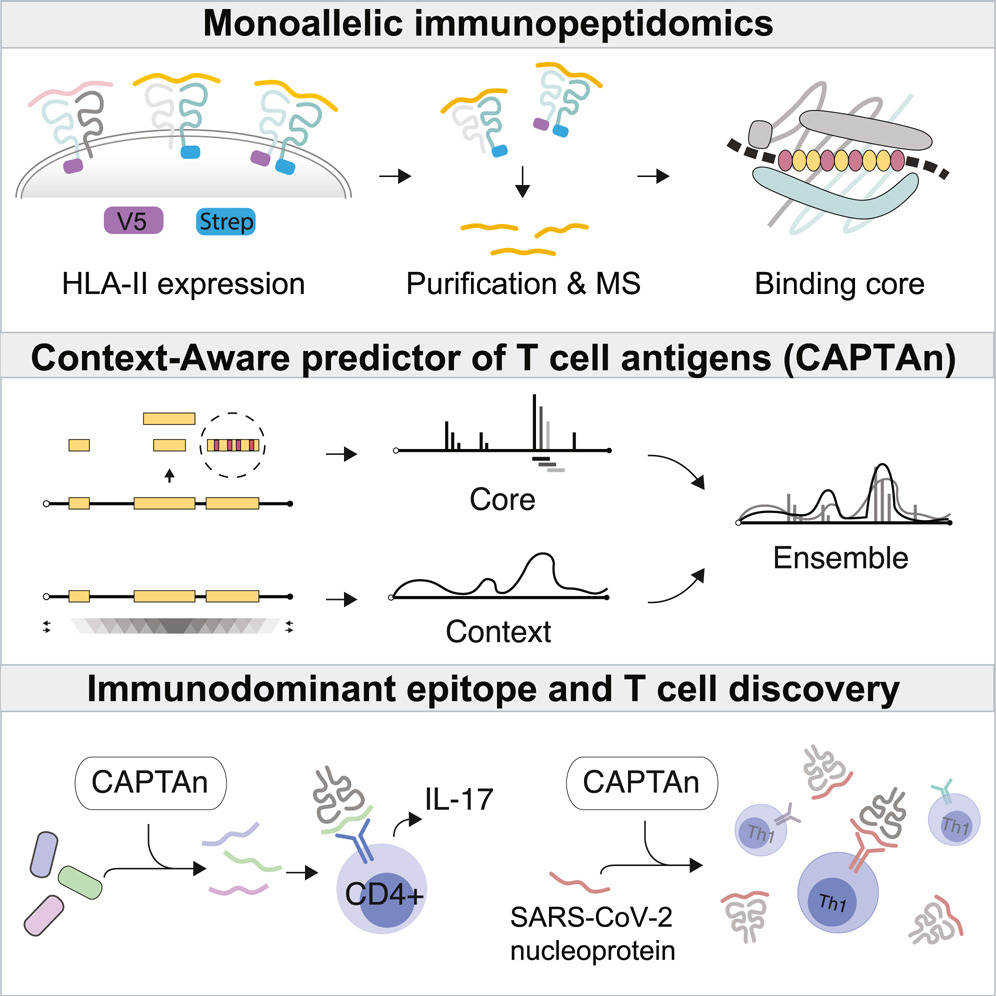
HLA-II immunopeptidome profiling and deep learning reveal features of antigenicity to inform antigen discovery.
Immunity. 2023;56(7):1681-1698.e13
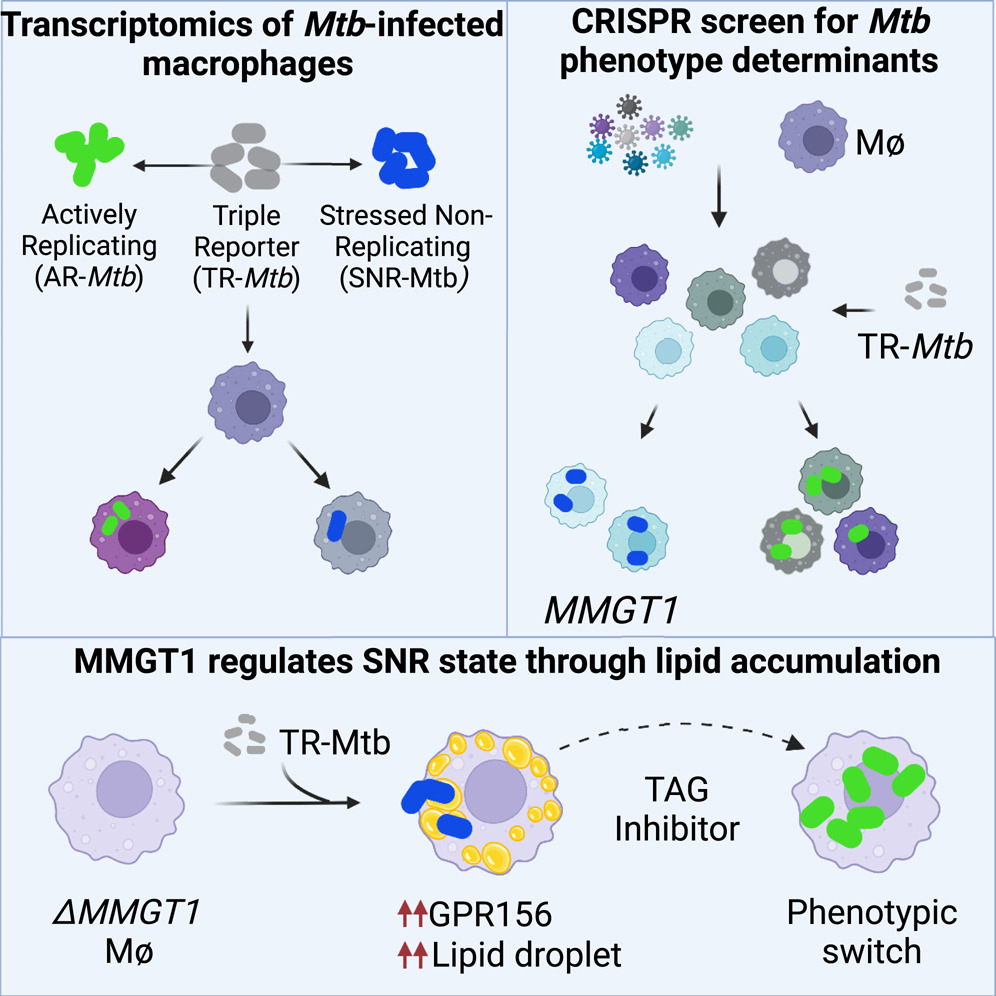
Identification of host regulators of Mycobacterium tuberculosis phenotypes uncovers a role for the MMGT1-GPR156 lipid droplet axis in persistence.
Cell Host Microbe. 2023;31(6):978-992.e5
Conditioning of the immune system by the microbiome.
Trends Immunol. 2023;44(7):499-511
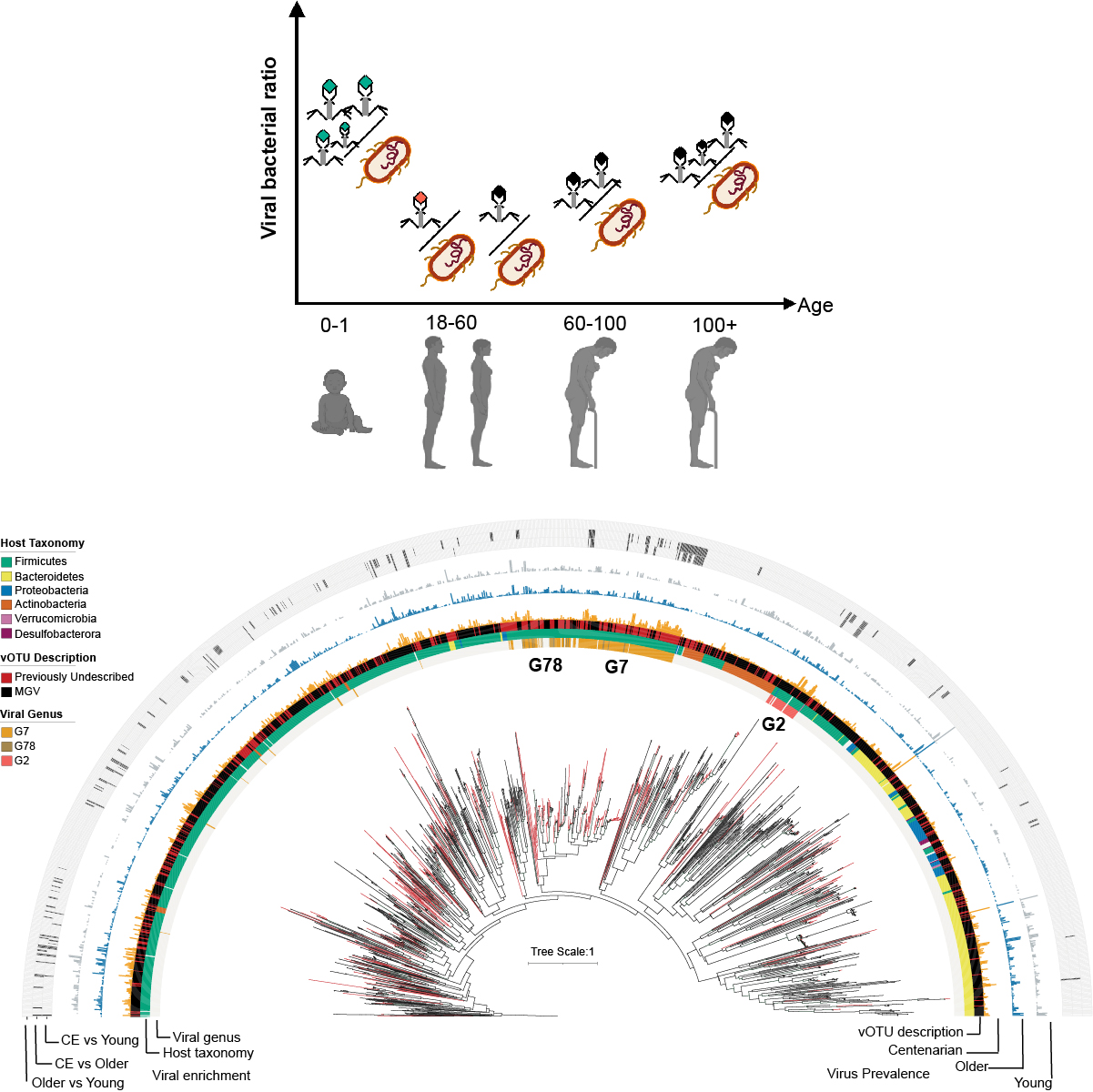
Centenarians have a diverse gut virome with the potential to modulate metabolism and promote healthy lifespan.
Nat Microbiol. 2023;8(6):1064-1078
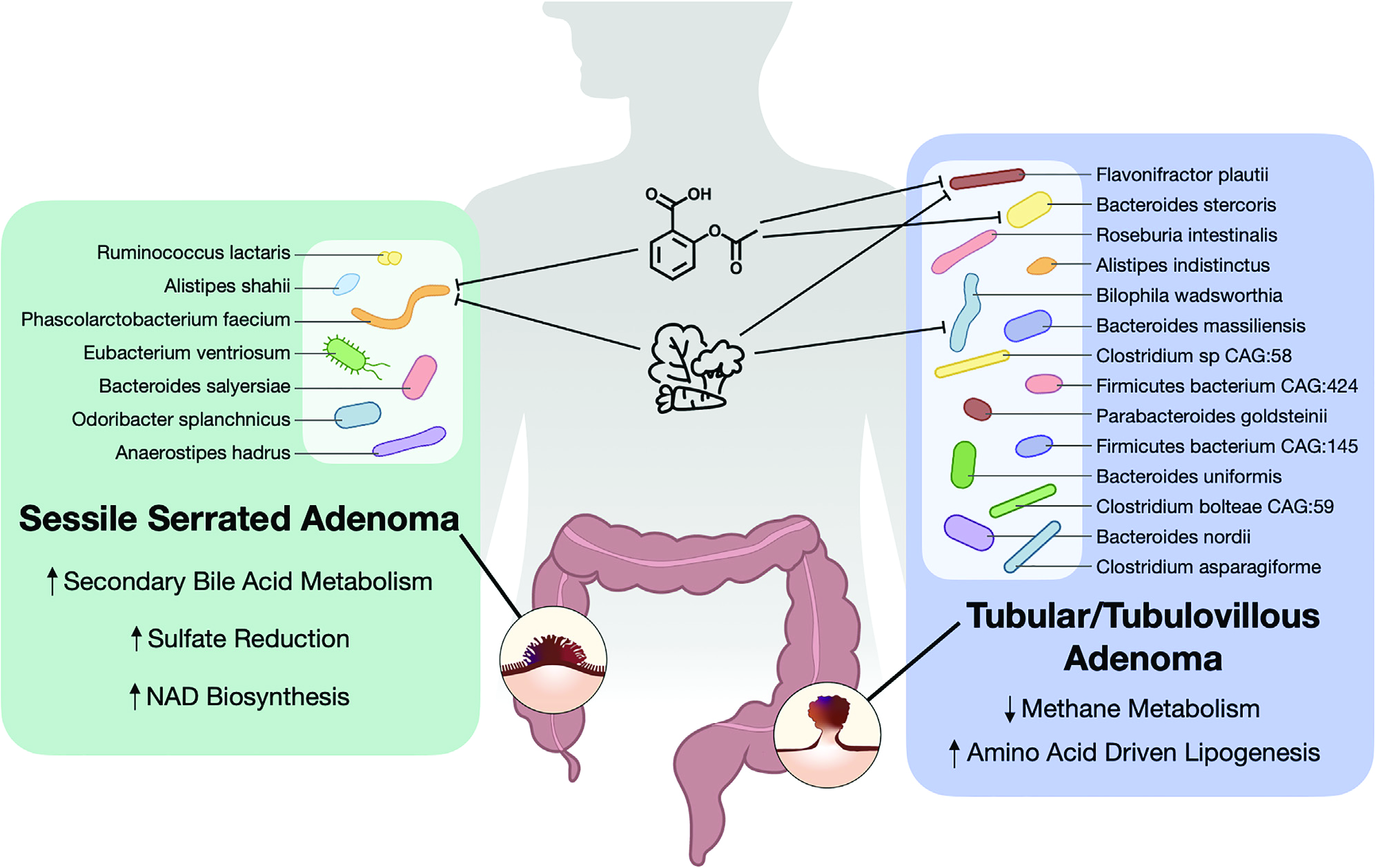
Association of distinct microbial signatures with premalignant colorectal adenomas.
Cell Host Microbe. 2023;31(5):827-838.e3
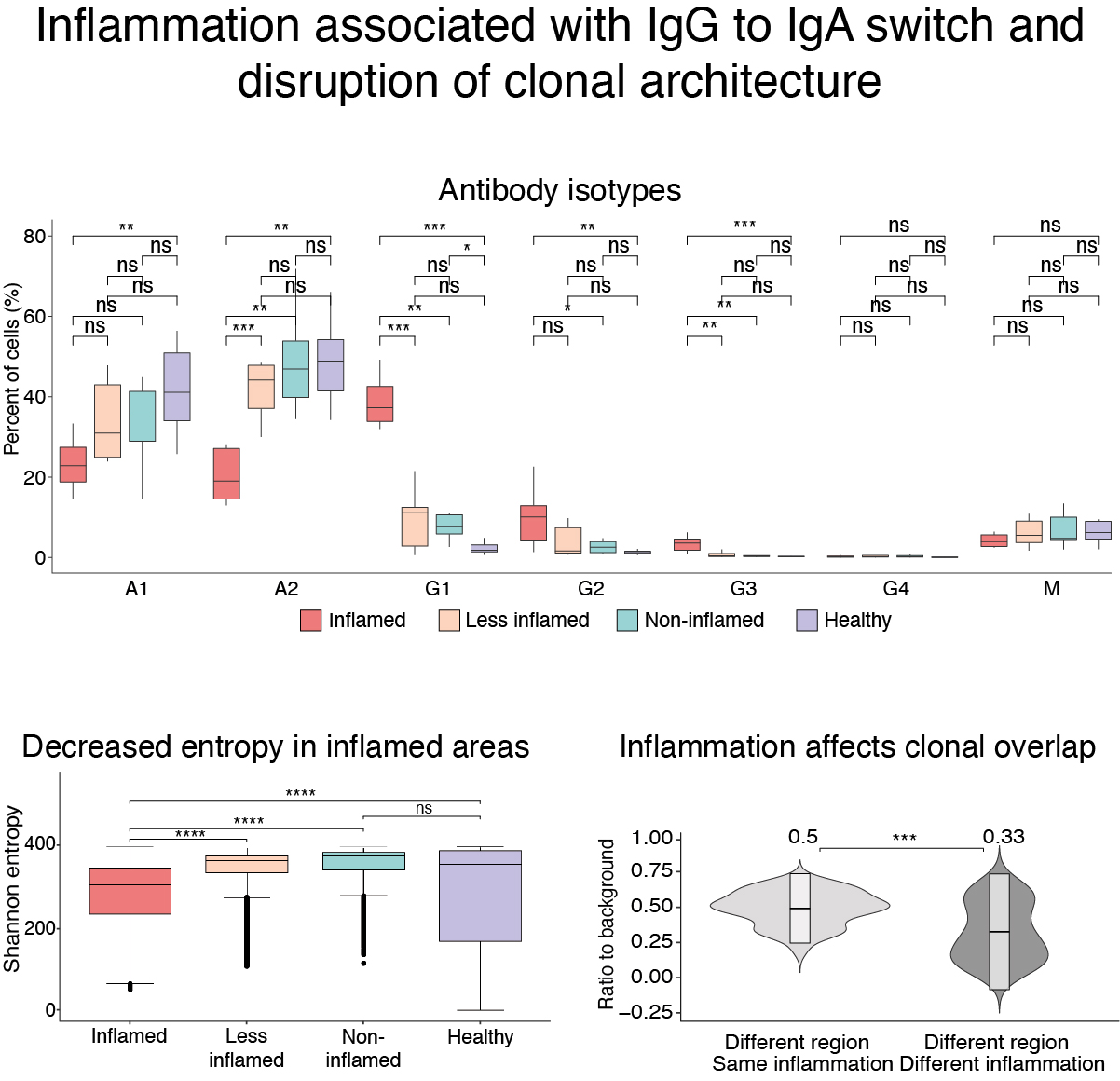
Remodeling of colon plasma cell repertoire within ulcerative colitis patients.
J Exp Med. 2023;220(4):ePub
Gut microbiome lipid metabolism and its impact on host physiology.
Cell Host Microbe. 2023;31(2):173-186