Quick Links
Labs
Xavier Lab
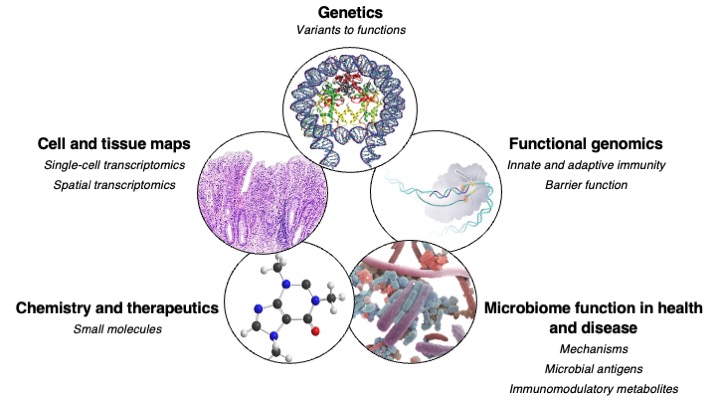
The Xavier Laboratory at the Massachusetts General Hospital and Broad Institute focuses on systematic characterization of genetic variants to understand the regulation of barrier defense, innate and adaptive immunity; chemical biology to control cellular disease phenotypes suggested by human genetics; molecular mechanisms to determine roles of the microbiome in health and disease; and development of computational approaches to uncover patterns of human and microbial pathway regulation during disease and treatment.