Quick Links
CAREER OPPORTUNITIES
We are always looking for talented post-docs, students, and research associates to join our team. Recent papers include:
Cell. 2022;185(26):4921-4936.e15
Nat Microbiol. 2022;7(10):1673-1685
Immunity. 2022;55(10):1909-1923.e6
Nature. 2022;608(7921):168-173
Nat Immunol. 2022;23(7):1063-1075
Cell. 2021;184(12):3205-3221.e24
Please contact us at xavier@molbio.mgh.harvard.edu
NEWS AND EVENTS
A spatial map of stricturing Crohn's disease. Fibrotic strictures are a major complication of Crohn’s disease (CD) that impair intestinal function, with no treatment aside from surgery. To address our limited understanding of the underlying mechanisms, we and the Smillie lab generated a single-cell and spatial transcriptomic atlas of stricturing CD. We found that intestinal strictures were characterized by increased immune cells, including IgG+ plasma cells, CCR7-hi CD4+ T cells, and inflammatory fibroblasts. We also mapped the expression of genetic risk variants onto intestinal biogeography, revealing that risk loci are enriched within discrete spatial niches defined by the presence of inflammatory fibroblasts and lymphoid follicles. Read more in Nature Genetics and a Broad news story.
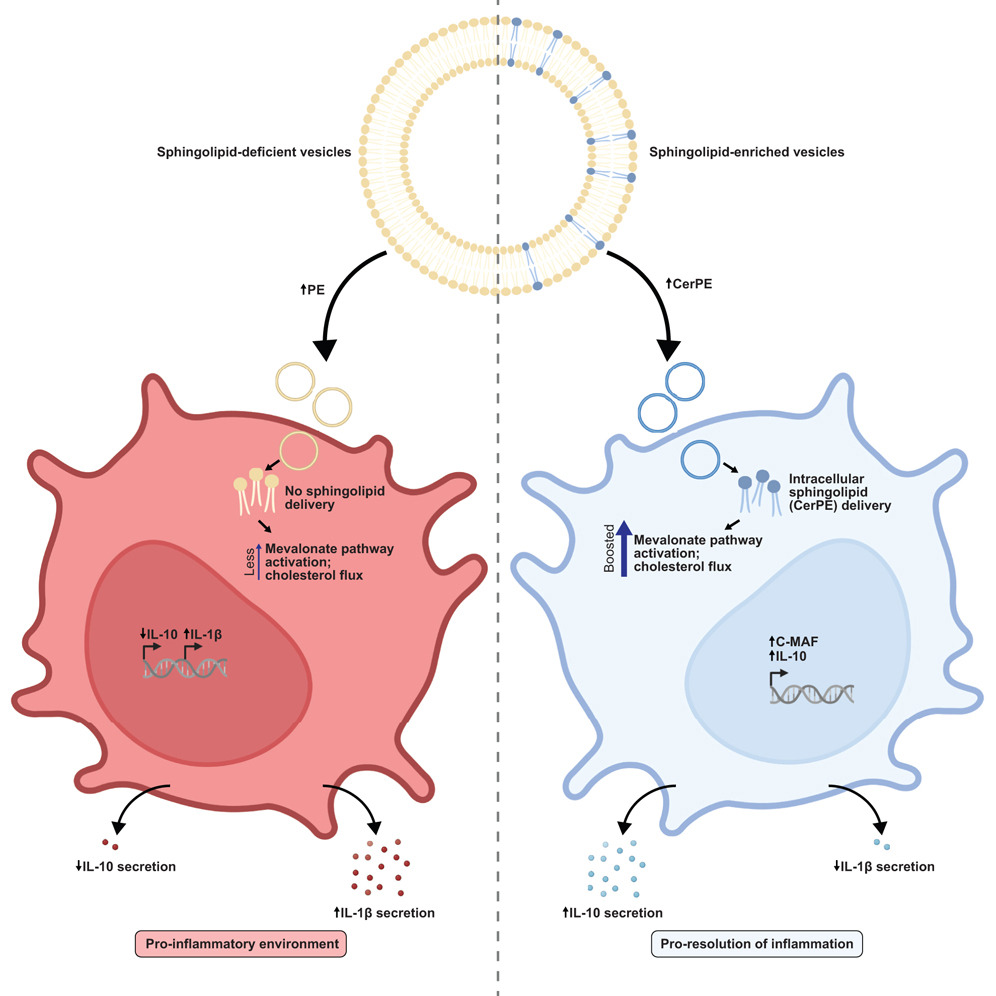 Bacteroides sphingolipids promote anti-inflammatory responses through the mevalonate pathway. Sphingolipids derived from ubiquitous gut Bacteroides species are associated with changes in host inflammation and metabolic syndrome; however, the signaling mechanisms within host cells are unknown. We utilized outer membrane vesicles (OMVs) from wild-type and sphingolipid-deficient Bacteroides strains to understand how these lipids modulate host inflammation. OMVs secreted by Bacteroides delivered sphingolipids into host dendritic cells, inducing an IL-10 response that is dependent on mevalonate pathway activation. Read more in Cell Host & Microbe.
Bacteroides sphingolipids promote anti-inflammatory responses through the mevalonate pathway. Sphingolipids derived from ubiquitous gut Bacteroides species are associated with changes in host inflammation and metabolic syndrome; however, the signaling mechanisms within host cells are unknown. We utilized outer membrane vesicles (OMVs) from wild-type and sphingolipid-deficient Bacteroides strains to understand how these lipids modulate host inflammation. OMVs secreted by Bacteroides delivered sphingolipids into host dendritic cells, inducing an IL-10 response that is dependent on mevalonate pathway activation. Read more in Cell Host & Microbe.
New clues to the role of neuro-immune crosstalk in gut infection. The enteric nervous system not only mediates digestion but also influences neuro-immune communication during gut infection in ways that are not yet fully understood. We collaborated with Vijay Kuchroo's group to explore the responses of enteric neurons to inflammatory stimuli using an animal model infected with the tissue-invading helminth Heligmosomoides polygyrus. Our study, published in Science, showed that primary enteric sensory neurons sense type 2 cytokines, govern a delicate balance of neuropeptides, and influence site-specific immunity.
 Taking aim at disease genes with a library of diverse molecular glues. In collaboration with Stuart Schreiber's group, we built a large and diverse collection of molecular compounds designed to bring two proteins together to target genetic variants linked to human disease in a new way. We use one protein, FKBP12, to stabilize another and reverse its pathological effects. We identified a compound that targets and reduces the detrimental effects of the Crohn's disease variant protein ATG16L1 T300A.This approach could be applied to recruit proteins with other functional effects or to engage targets in specific cell types. Read more in Cell Chemical Biology and a Broad news story.
Taking aim at disease genes with a library of diverse molecular glues. In collaboration with Stuart Schreiber's group, we built a large and diverse collection of molecular compounds designed to bring two proteins together to target genetic variants linked to human disease in a new way. We use one protein, FKBP12, to stabilize another and reverse its pathological effects. We identified a compound that targets and reduces the detrimental effects of the Crohn's disease variant protein ATG16L1 T300A.This approach could be applied to recruit proteins with other functional effects or to engage targets in specific cell types. Read more in Cell Chemical Biology and a Broad news story.
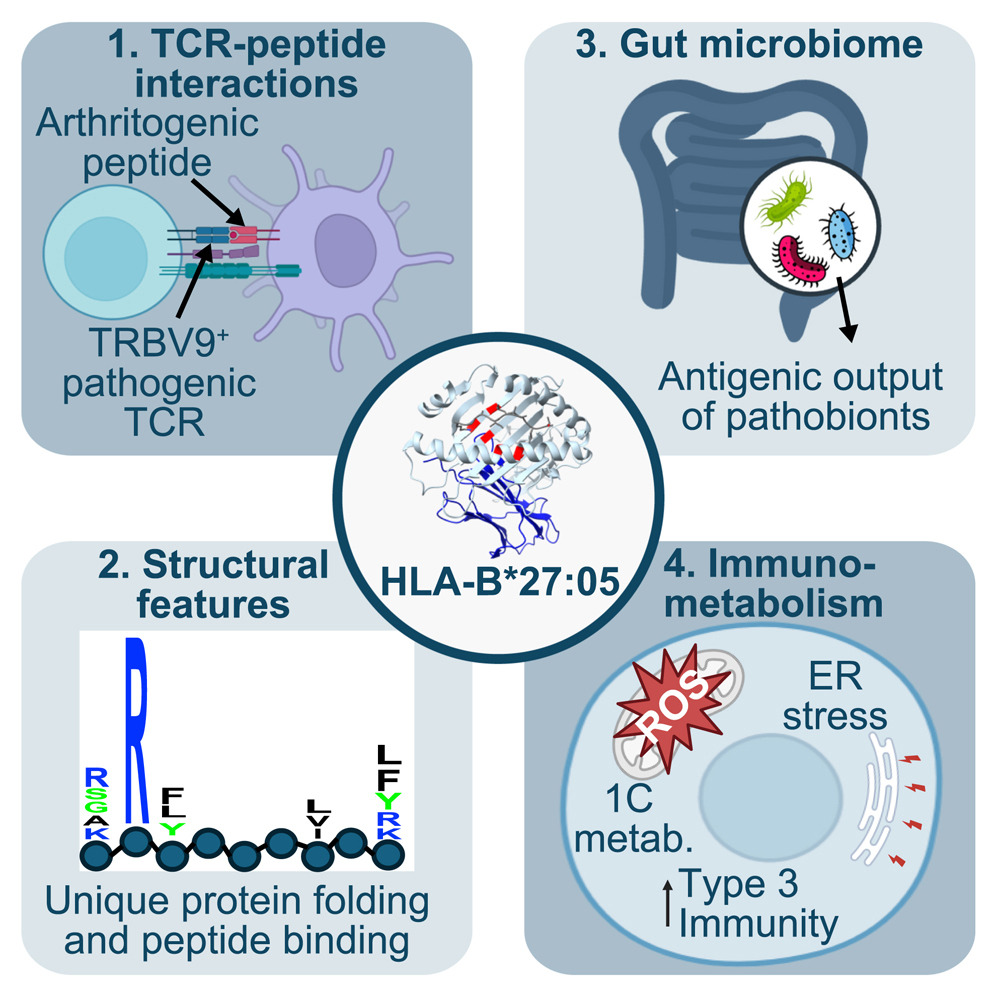 Why HLA-B27 confers risk for spondyloarthritis: biochemical, microbial, and immunological evidence. The strong association of human leukocyte antigen B27 alleles (HLA-B27) with spondyloarthritis and related rheumatic conditions has long fascinated researchers, yet the precise mechanisms underlying their pathogenicity remain elusive. In Cell Chemical Biology, we reviewed how interplay between the microbiome, the immune system, and HLA-B27 could trigger spondyloarthritis, with a focus on whether HLA-B27 presents an arthritogenic peptide.
Why HLA-B27 confers risk for spondyloarthritis: biochemical, microbial, and immunological evidence. The strong association of human leukocyte antigen B27 alleles (HLA-B27) with spondyloarthritis and related rheumatic conditions has long fascinated researchers, yet the precise mechanisms underlying their pathogenicity remain elusive. In Cell Chemical Biology, we reviewed how interplay between the microbiome, the immune system, and HLA-B27 could trigger spondyloarthritis, with a focus on whether HLA-B27 presents an arthritogenic peptide.
 On the Cover – Microbial metabolites mediate host-microbiome communication. The cover art for our latest Cell Host & Microbe article illustrates how we linked disease-associated microbes with changes in gut and plasma metabolites in a pediatric ulcerative colitis cohort to uncover host-microbial interactions that contribute to inflammatory pathology. Congratulations to Melanie Schirmer (now faculty at the Technical University of Munich), Martin Stražar, and colleagues on this paper and its amazing cover!
On the Cover – Microbial metabolites mediate host-microbiome communication. The cover art for our latest Cell Host & Microbe article illustrates how we linked disease-associated microbes with changes in gut and plasma metabolites in a pediatric ulcerative colitis cohort to uncover host-microbial interactions that contribute to inflammatory pathology. Congratulations to Melanie Schirmer (now faculty at the Technical University of Munich), Martin Stražar, and colleagues on this paper and its amazing cover!
The Gene Lay Institute of Immunology and Inflammation established. On May 9, Brigham and Women’s Hospital announced that a momentous $100 million gift from BioLegend founder and CEO Gene Lay, MS, DVM has established The Gene Lay Institute of Immunology and Inflammation. This gift opens a promising new chapter of better understanding the roles that immunology and inflammation play in disease. Vijay Kuchroo, DVM, PhD will serve as director of this new institute along with vice directors Arlene Sharpe, MD, PhD and CCIB's Ramnik Xavier, MD, PhD. Learn more about the institute here.
In the News – Tiny genetic sequences in a mother's bacteria seem to hop into the infant's bacteria, perhaps ensuring a healthy microbiome later in life. Quanta magazine reviews our recent Vatanen et al. Cell paper. You can read their full article here.
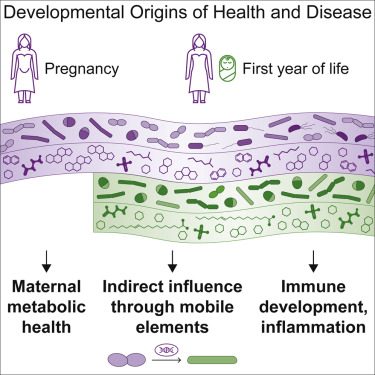 Congrats go two-fold to Tommi Vatanen! Tommi was named a Highly Cited Researcher 2022 by @ClarivateAG and is the first author of a Cell paper detailing how a mother’s microbiome helps shape her baby’s development. In the study, Tommi and collaborators in the Xavier lab and beyond, tracked the co-development of microbiomes and metabolomes in a cohort of 70 mother–infant pairs from late pregnancy to 1 year of age. They uncovered an additional mode of vertical transmission, where maternal gut bacteria share genes with infant gut strains in the absence of stable engraftment of the maternal bacteria themselves. The genes were frequently involved in functions related to metabolism of infant dietary substrates. The study expands our understanding of maternal influences on the infant gut microbiome, with potentially profound implications for immune and neurodevelopment early in life.
Congrats go two-fold to Tommi Vatanen! Tommi was named a Highly Cited Researcher 2022 by @ClarivateAG and is the first author of a Cell paper detailing how a mother’s microbiome helps shape her baby’s development. In the study, Tommi and collaborators in the Xavier lab and beyond, tracked the co-development of microbiomes and metabolomes in a cohort of 70 mother–infant pairs from late pregnancy to 1 year of age. They uncovered an additional mode of vertical transmission, where maternal gut bacteria share genes with infant gut strains in the absence of stable engraftment of the maternal bacteria themselves. The genes were frequently involved in functions related to metabolism of infant dietary substrates. The study expands our understanding of maternal influences on the infant gut microbiome, with potentially profound implications for immune and neurodevelopment early in life.
Lipid immunogens – a key trigger for the inflammatory immune response. In collaboration with the Clardy lab at Harvard Medical School, we report in JACS a small but critical correction to the structure of a lipid immunogen for Lyme Disease, with implications for vaccine efforts and immune activation by bacterial galactolipids.
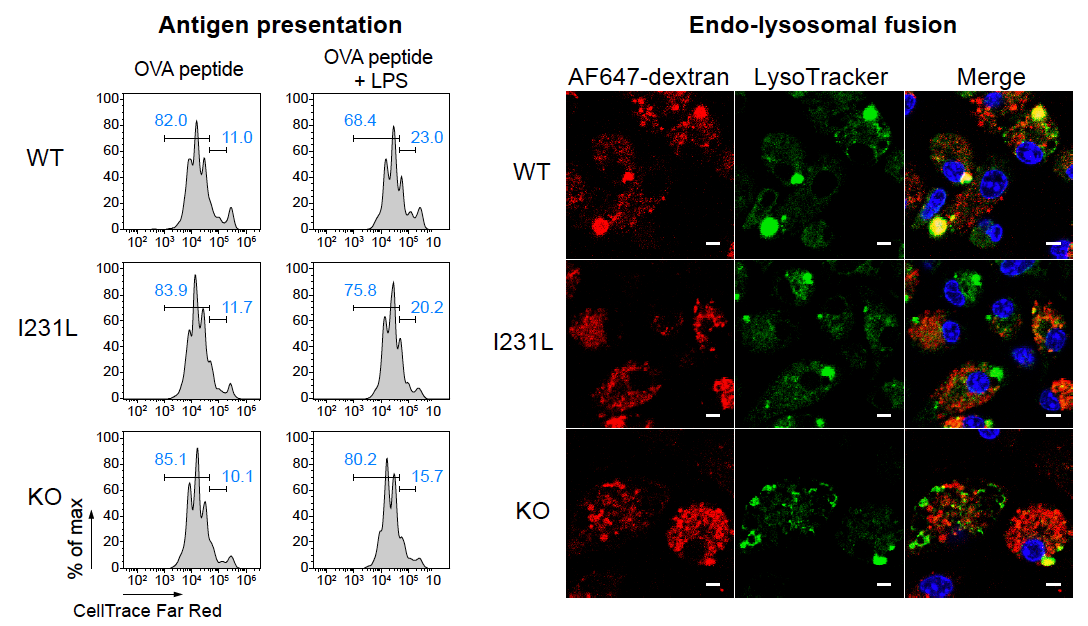 Congrats to Xiangjun Chen! and colleagues on identifying pH sensor GPR65 as a controller of tissue inflammation in vivo and revealing functions of the GPR65 I231L inflammatory bowel disease risk variant in multiple immune cell populations. Mice carrying this variant had impaired antibacterial defenses in an infection model and heightened inflammation in a colitis model. GPR65 I231L led to cytokine imbalance and altered metabolism, and increased the release of IL2 and IL3 in dendritic cells at acidic pH, leading to enhanced antigen presentation. Read more in Nature Immunology.
Congrats to Xiangjun Chen! and colleagues on identifying pH sensor GPR65 as a controller of tissue inflammation in vivo and revealing functions of the GPR65 I231L inflammatory bowel disease risk variant in multiple immune cell populations. Mice carrying this variant had impaired antibacterial defenses in an infection model and heightened inflammation in a colitis model. GPR65 I231L led to cytokine imbalance and altered metabolism, and increased the release of IL2 and IL3 in dendritic cells at acidic pH, leading to enhanced antigen presentation. Read more in Nature Immunology.
Large scale sequencing efforts link new genes to Crohn's disease risk. To better understand the biology of Crohn's disease, the International Inflammatory Bowel Disease Genetics Consortium sequenced the protein-coding exomes of more than 100,000 people from 12 countries. They identified 10 genes that affect a person's risk of Crohn's disease, six of which have never before been linked to the disorder. Learn more in Nature Genetics.
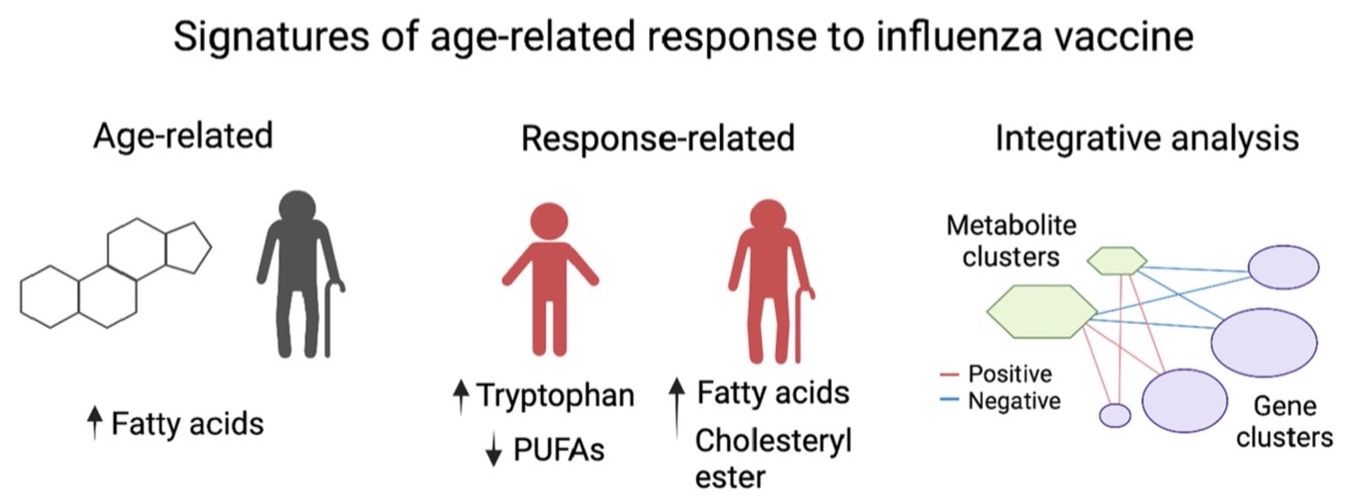 Congrats to Chih-Hung Chou! and colleagues on a collaborative study with Albert Shaw's lab at Yale University that uncovered age-related metabolomic and transcriptomic signatures associated with influenza vaccine response. Differences in response between age groups was associated with signatures derived from metabolic pathways involved in fatty acid and sterol lipid metabolism. Young high responders showed increased tryptophan pathway metabolites and reduced levels of polyunsaturated fatty acids. In contrast, older high responders show increased fatty acid synthesis and accumulation of cholesteryl esters. Uncovering these molecular profiles will help to identify populations at risk of influenza vaccine failure, as well as to inform design of vaccine development trials to improve vaccine efficacy. You can read the full study in Aging Cell.
Congrats to Chih-Hung Chou! and colleagues on a collaborative study with Albert Shaw's lab at Yale University that uncovered age-related metabolomic and transcriptomic signatures associated with influenza vaccine response. Differences in response between age groups was associated with signatures derived from metabolic pathways involved in fatty acid and sterol lipid metabolism. Young high responders showed increased tryptophan pathway metabolites and reduced levels of polyunsaturated fatty acids. In contrast, older high responders show increased fatty acid synthesis and accumulation of cholesteryl esters. Uncovering these molecular profiles will help to identify populations at risk of influenza vaccine failure, as well as to inform design of vaccine development trials to improve vaccine efficacy. You can read the full study in Aging Cell.
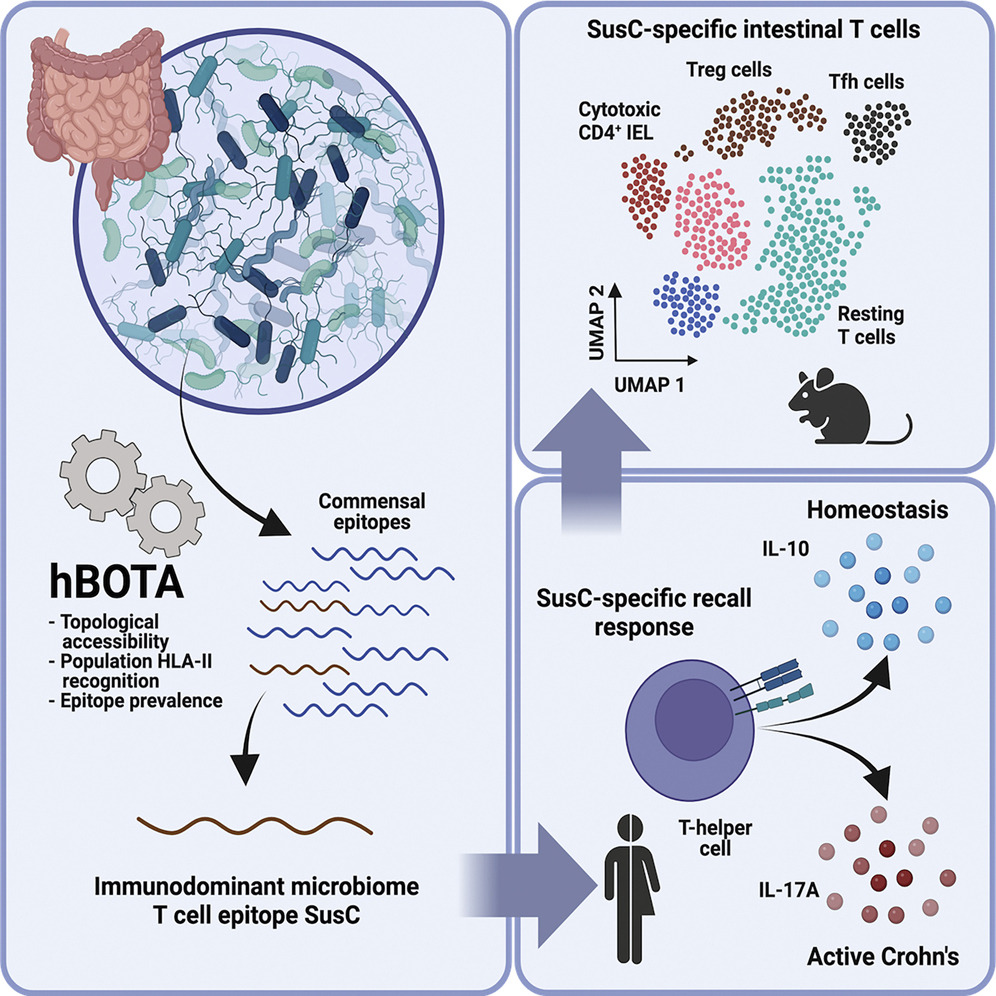 Identifying helpful bacteria. The gut microbiome plays a vital role in creating a healthy gastrointestinal environment. So how do the body's natural defenses tell helpful and harmful bacteria apart? We used a computational algorithm to look for antigen-coding DNA sequences in microbial genes and identified several antigens that were recognized — but tolerated — by the immune system. Our findings suggest that some of these antigens could one day be used to monitor inflammatory bowel diseases.
Identifying helpful bacteria. The gut microbiome plays a vital role in creating a healthy gastrointestinal environment. So how do the body's natural defenses tell helpful and harmful bacteria apart? We used a computational algorithm to look for antigen-coding DNA sequences in microbial genes and identified several antigens that were recognized — but tolerated — by the immune system. Our findings suggest that some of these antigens could one day be used to monitor inflammatory bowel diseases.
How oral bacteria build a new home in the gut. Oral bacteria in the gut can cause inflammatory bowel disease (IBD), cancer, and other disorders. To understand how they colonize the intestines, we studied Veillonella parvula, an oral microbe that grows in the guts of IBD patients. We found that the bacteria use nitrate, a metabolite of inflammation, to switch to anaerobic respiration and also turn to organic acids, amino acids, and peptides as carbon sources, fueling its growth. Nitrate respiration allowed the microbe to colonize the gut in a mouse model of colitis. The team concludes that V. parvula takes advantage of inflammatory conditions to colonize. Read more in Nature Microbiology.
The gut microbiome: from associations to function. In collaboration with the Clardy lab, the Xavier lab has identified a lipid from the Akkermansia muciniphila cell membrane that functions in homeostatic immunity. The gut microbe A. muciniphila has been strongly associated with positive systemic effects on host metabolism and favorable immune checkpoint blockade outcomes for cancer. The study, published in Nature, explores the molecular mechanism underlying these associations. The authors identified a novel lipid from the bacteria’s cell membrane, determined its molecular structure, and discovered that it binds to two toll-like receptors on immune cells, triggering the release of certain immune-stimulating cytokines. The team also revealed how a single molecule and microbial chemistry help moderate immune responses, and suggest their findings could inspire new therapeutics. Read more in this HMS news story.
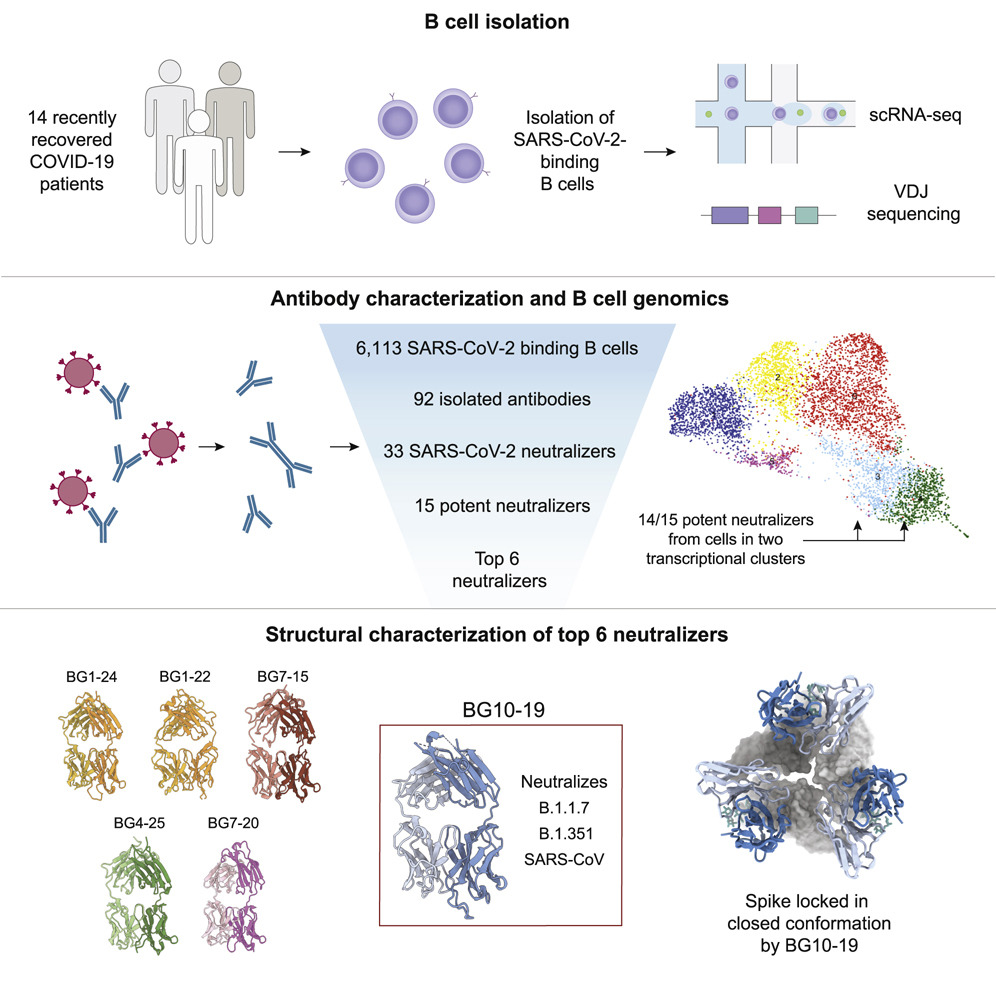 Why are certain B cells more effective against SARS-CoV-2? Using single-cell RNA sequencing and other methods to study blood samples from 14 people who recovered from COVID-19, Johannes Scheid, Basak Eraslan and collaborators identified distinct patterns of gene expression in B cells that produce antibodies that bind tightly to and neutralize SARS-CoV-2. They also discovered a new antibody, BG10-19, that neutralizes SARS-CoV2 and the coronavirus that caused the 2003 SARS outbreak. Their findings, published in Cell, suggest some B cells are especially effective at producing antibodies, and offer insight on how to design antibodies against several coronaviruses.
Why are certain B cells more effective against SARS-CoV-2? Using single-cell RNA sequencing and other methods to study blood samples from 14 people who recovered from COVID-19, Johannes Scheid, Basak Eraslan and collaborators identified distinct patterns of gene expression in B cells that produce antibodies that bind tightly to and neutralize SARS-CoV-2. They also discovered a new antibody, BG10-19, that neutralizes SARS-CoV2 and the coronavirus that caused the 2003 SARS outbreak. Their findings, published in Cell, suggest some B cells are especially effective at producing antibodies, and offer insight on how to design antibodies against several coronaviruses.
A unique community of gut microbes in centenarians may help support longevity. In a collaboration with Kenya Honda’s lab to characterize gut microbiomes of 160 Japanese centenarians, the Xavier lab found that, compared to younger people, the centenarians showed higher levels of intestinal microbes that produce secondary bile acids such as isoalloLCA, which strongly inhibits the growth of some gram-positive bacteria - see the news story. Their findings, published in Nature, could help researchers uncover mechanisms and biomarkers of healthy aging, and develop new ways to treat chronic inflammation and bacterial disease.
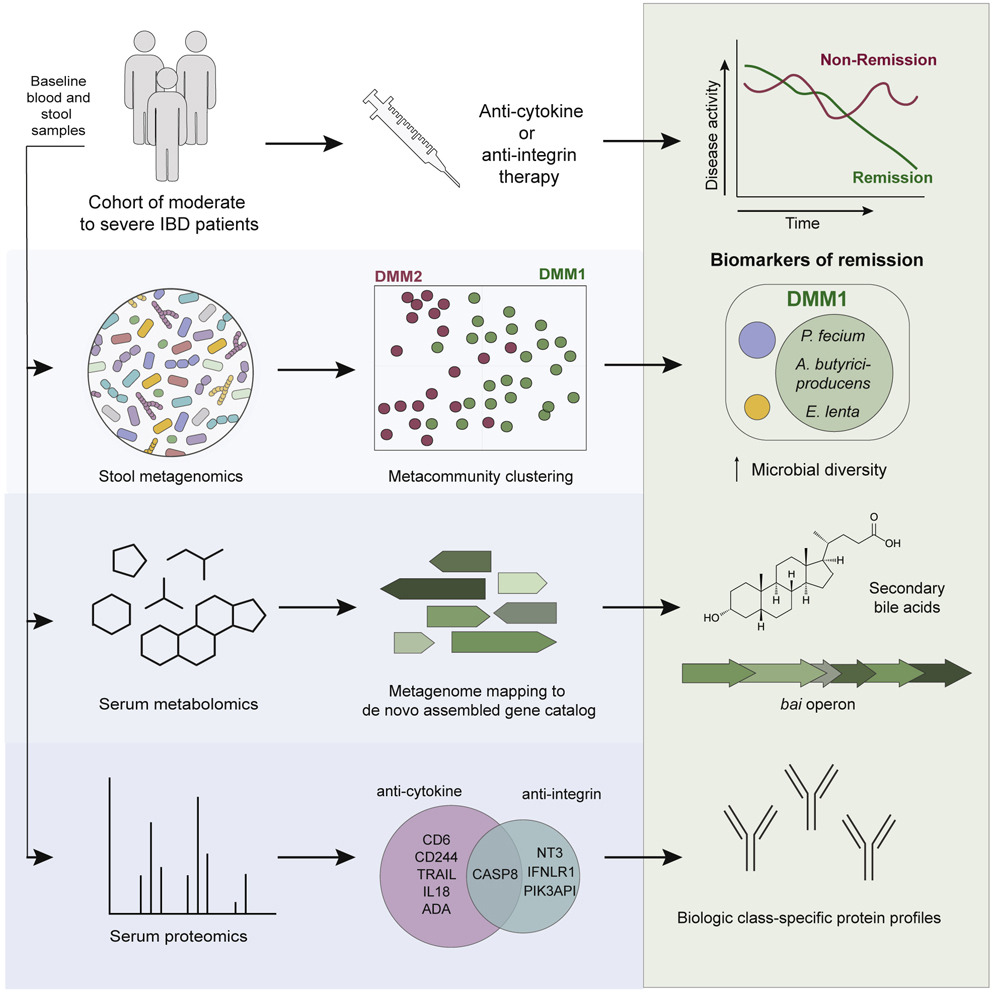 Identifying multiomic markers of IBD treatment response. It is difficult to predict response to treatment for inflammatory bowel disease (IBD) because of the complexity of the intestinal microbiome and the lack of predictive biomarkers. In a study published in Cell Host & Microbe, Jonathan Wei Jie Lee and Damian Plichta in the Xavier lab and colleagues profiled stool and blood samples from patients with IBD before treatment and tracked treatment response. They used modeling and machine learning to identify metagenomic, metabolomic, and proteomic markers that could predict which patients would achieve remission. Biomarkers varied by therapy class, with microbial diversity in the gut being a strong indicator of anti-cytokine therapy success. These response markers could lead to better IBD treatment selection.
Identifying multiomic markers of IBD treatment response. It is difficult to predict response to treatment for inflammatory bowel disease (IBD) because of the complexity of the intestinal microbiome and the lack of predictive biomarkers. In a study published in Cell Host & Microbe, Jonathan Wei Jie Lee and Damian Plichta in the Xavier lab and colleagues profiled stool and blood samples from patients with IBD before treatment and tracked treatment response. They used modeling and machine learning to identify metagenomic, metabolomic, and proteomic markers that could predict which patients would achieve remission. Biomarkers varied by therapy class, with microbial diversity in the gut being a strong indicator of anti-cytokine therapy success. These response markers could lead to better IBD treatment selection.
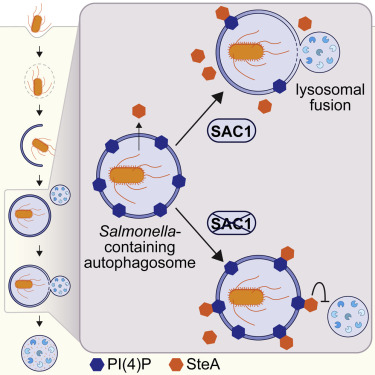 Despite the importance of membrane identity and trafficking in selective autophagy, the role of lipid membrane composition remains largely unknown. In a study out in Cell Reports, Kai Liu and colleagues in the Xavier lab identified the PI(4)P phosphatase SAC1 as an essential regulator of xenophagy during Salmonella infection. Loss of SAC1 activity delays fusion between Salmonella-containing autophagosomes and lysosomes, leading to increased intracellular bacterial replication. They additionally found that the Salmonella type III secreted effector and PI(4)P-binding protein, SteA, promotes replication by impeding lysosomal fusion.
Despite the importance of membrane identity and trafficking in selective autophagy, the role of lipid membrane composition remains largely unknown. In a study out in Cell Reports, Kai Liu and colleagues in the Xavier lab identified the PI(4)P phosphatase SAC1 as an essential regulator of xenophagy during Salmonella infection. Loss of SAC1 activity delays fusion between Salmonella-containing autophagosomes and lysosomes, leading to increased intracellular bacterial replication. They additionally found that the Salmonella type III secreted effector and PI(4)P-binding protein, SteA, promotes replication by impeding lysosomal fusion.
Type 1 diabetes could become a preventable disease pending clinical trials with B. infantis. An upcoming clinical study will test whether an @EvolveBio B. infantis probiotic can prevent the development of type 1 diabetes in genetically predisposed infants - see the news story. DIABIMMUNE was one of the key studies that clearly associated gut microbiota with the development of type 1 diabetes. In this study, scientists collected stool samples from children living in Finland, Russian Karelia, and Estonia from birth to 3 years of age. Though they share a border, children in Finland and Russian Karellia grow up in very different environments. Higher levels of modernization in Finland coincide with higher incidences of type 1 diabetes and other autoimmune diseases. The stark contrasts in the microbiota of the DIABIMMUNE children was first reported by Tommi Vatanen, a computational biologist and microbiologist in the Xavier lab.
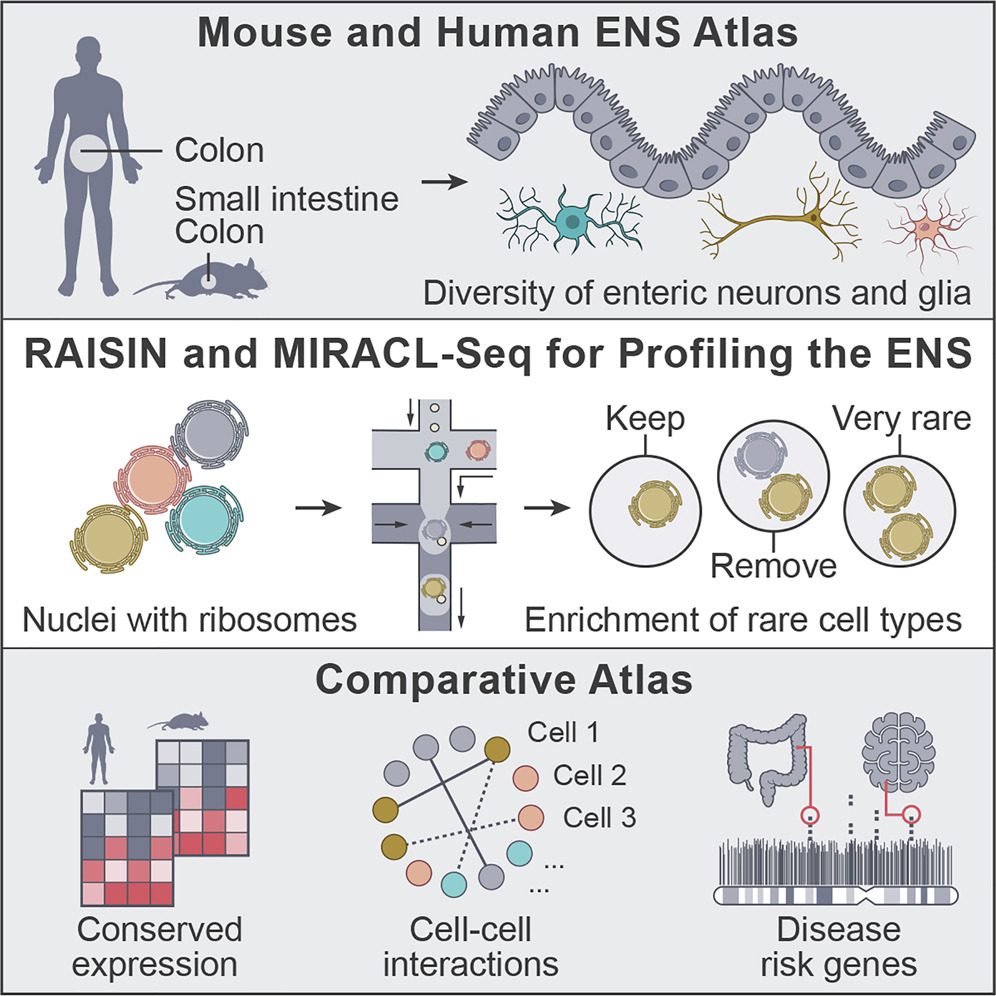 The enteric nervous system as a hub for intestinal and immune cell crosstalk. Studying enteric neurons is a challenge because they are rare, fragile, and not easy to isolate from surrounding tissue. A team led by Chris Smillie, Eugene Drokhlyansky, Ramnik Xavier and Aviv Regev devised two new methods to study the enteric nervous system (ENS) in mice and humans at single-cell resolution: RAISIN RNA-seq and MIRACL-seq. Together, these techniques enabled the team to infer that neurons in the gut are communicating with a variety of other cell types, including immune cells. They also found that key genes associated with disease are expressed in the ENS. Read more in the full article in Cell.
The enteric nervous system as a hub for intestinal and immune cell crosstalk. Studying enteric neurons is a challenge because they are rare, fragile, and not easy to isolate from surrounding tissue. A team led by Chris Smillie, Eugene Drokhlyansky, Ramnik Xavier and Aviv Regev devised two new methods to study the enteric nervous system (ENS) in mice and humans at single-cell resolution: RAISIN RNA-seq and MIRACL-seq. Together, these techniques enabled the team to infer that neurons in the gut are communicating with a variety of other cell types, including immune cells. They also found that key genes associated with disease are expressed in the ENS. Read more in the full article in Cell.
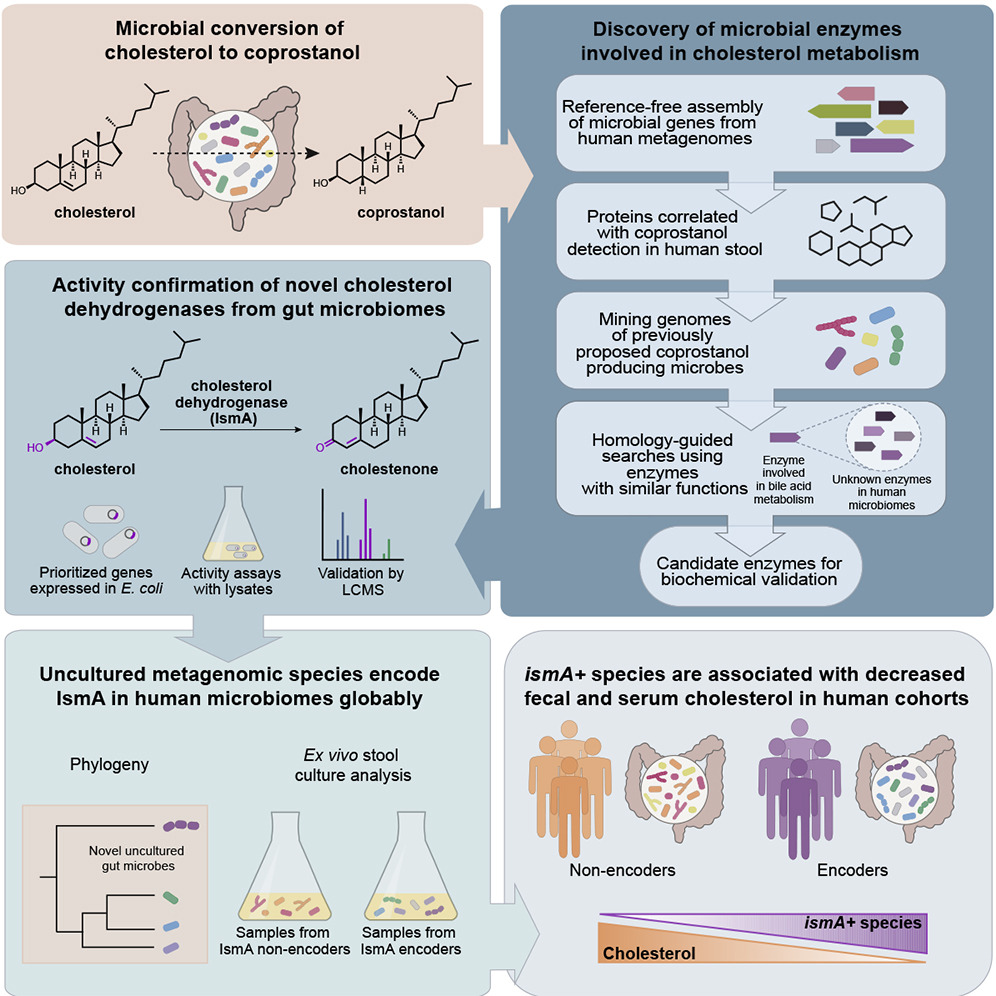 Gut bacteria that metabolize cholesterol may have a positive impact on people's cardiac health. In a study published in Cell Host & Microbe, Doug Kenny in the Xavier lab and colleagues found that people who have cholesterol metabolizing bacteria in their intestines have lower cholesterol levels in their blood than those without the microbes. The discovery suggests a possible reason why some people can consume more cholesterol in their diet with minimal effect on their blood cholesterol levels. It also hints that boosting populations of these bacteria, through diet, probiotics, or another kind of treatment, may one day be an effective way to help lower cholesterol levels.
Gut bacteria that metabolize cholesterol may have a positive impact on people's cardiac health. In a study published in Cell Host & Microbe, Doug Kenny in the Xavier lab and colleagues found that people who have cholesterol metabolizing bacteria in their intestines have lower cholesterol levels in their blood than those without the microbes. The discovery suggests a possible reason why some people can consume more cholesterol in their diet with minimal effect on their blood cholesterol levels. It also hints that boosting populations of these bacteria, through diet, probiotics, or another kind of treatment, may one day be an effective way to help lower cholesterol levels.
Therapeutic opportunities in inflammatory bowel disease: mechanistic dissection of host-microbiome relationships. Mechanistic and functional links between host-microbiome interplay and irritable bowel disease pathology are building and presenting exciting but challenging opportunities to manipulate the microbiome for therapeutic benefit. A summary of the review is available here.
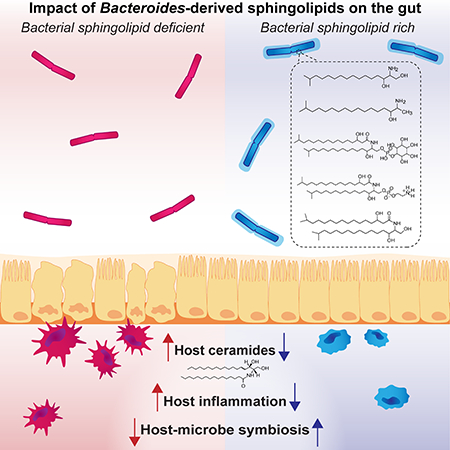 Gut sphingolipids send signals about health and disease. Sphingolipids, a signaling molecule and structural component of both bacterial and mammalian cell membranes, play a central role in regulating inflammation, immunity, growth, and cell survival. To date, the specific roles of bacterial sphingolipids in regulating innate immunity or metabolism in the mammalian gut have been largely unknown. Reporting in Cell Host & Microbe, Eric Brown, Ramnik Xavier, Hera Vlamakis, Clary Clish and colleagues describe sphingolipid metabolite alterations in stool as a defining signature in inflammatory bowel disease in humans and further demonstrate in mice that bacterial sphingolipid production plays a significant role in the host’s gut health and disease.
Gut sphingolipids send signals about health and disease. Sphingolipids, a signaling molecule and structural component of both bacterial and mammalian cell membranes, play a central role in regulating inflammation, immunity, growth, and cell survival. To date, the specific roles of bacterial sphingolipids in regulating innate immunity or metabolism in the mammalian gut have been largely unknown. Reporting in Cell Host & Microbe, Eric Brown, Ramnik Xavier, Hera Vlamakis, Clary Clish and colleagues describe sphingolipid metabolite alterations in stool as a defining signature in inflammatory bowel disease in humans and further demonstrate in mice that bacterial sphingolipid production plays a significant role in the host’s gut health and disease.
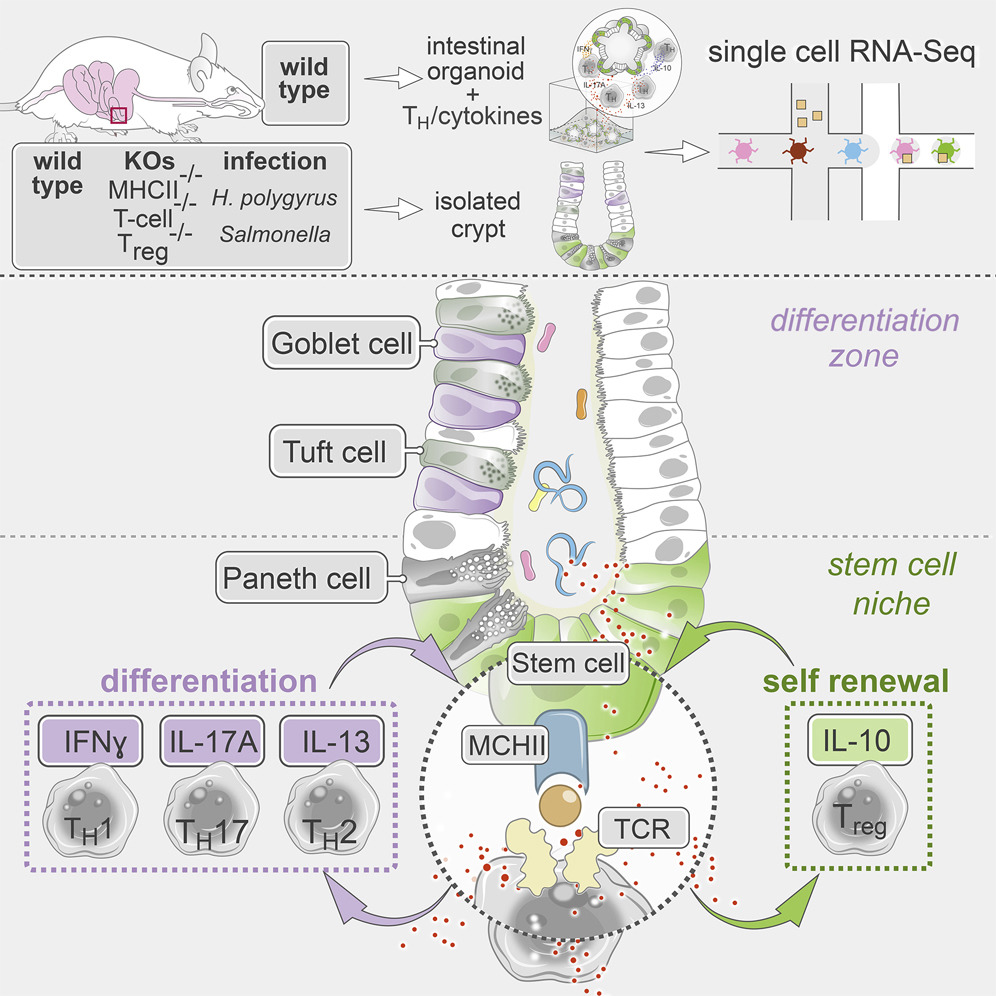 Eavesdropping on gut gossip. The gut's intestinal stem cells (ISCs) "talk" to other cell types to help maintain a robust and healthy cellular community. In this week's Cell, Moshe Biton, Adam Haber, Noga Rogel, Aviv Regev, Ramnik Xavier, and colleagues dive into one such conversation, between ISCs and T helper (Th) cells. They found that some ISCs express MCH II (a surface complex that activates Th cells), while Th cells produce cytokines (chemical messages) that influence ISCs’ behavior. The crosstalk may help maintain the right balance of immune activity in the gut, as well as a hardy stem cell pool. Read more in a Broad news story.
Eavesdropping on gut gossip. The gut's intestinal stem cells (ISCs) "talk" to other cell types to help maintain a robust and healthy cellular community. In this week's Cell, Moshe Biton, Adam Haber, Noga Rogel, Aviv Regev, Ramnik Xavier, and colleagues dive into one such conversation, between ISCs and T helper (Th) cells. They found that some ISCs express MCH II (a surface complex that activates Th cells), while Th cells produce cytokines (chemical messages) that influence ISCs’ behavior. The crosstalk may help maintain the right balance of immune activity in the gut, as well as a hardy stem cell pool. Read more in a Broad news story.
Peptide presentation prognosticator produced. We don't fully understand the immune system's rules for determining whether a peptide from a bacterium or other source will 1) be presented to T cells, and 2) spark an immune response. To help bring new clarity, Dan Graham, Chengwei Luo, Ramnik Xavier, and colleagues profiled the "peptidome" of potential antigens bound to MHC class II (a protein complex that presents peptides to T cells) in mice. They used their data to build and train BOTA, a machine learning algorithm that predicts antigenic peptides based on bacterial whole genome data. Learn more in Nature Medicine and in a Broad news story. A comprehensive summary of the study can be found here.


